Towards pharmacokinetic profile predictions for monoclonal antibodies using sequence based machine learning derived parameters and compartmental modeling
Towards pharmacokinetic profile predictions for monoclonal antibodies using sequence based machine learning derived parameters and compartmental modeling
Jost, F.; Cordes, H.
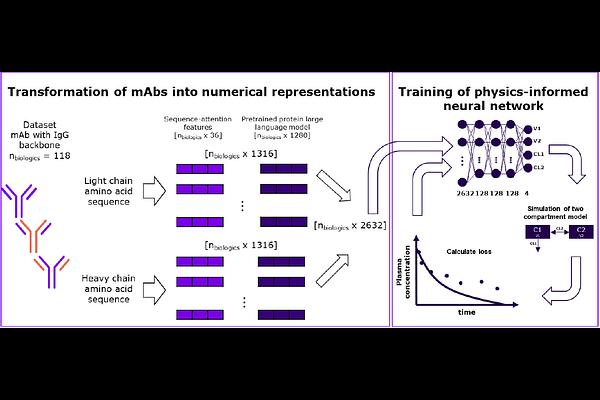
AbstractThis study presents a novel approach for predicting complete pharmacokinetic (PK) profiles of monoclonal antibodies (mAbs) based solely on their amino acid sequences, addressing a critical gap in early-stage therapeutic antibody development. While current methodologies rely on in vivo testing in specialized models like Tg32 mice or cynomolgus monkeys, our approach enables PK parameter prediction prior to molecule synthesis. Using a dataset of 118 diverse mAbs, we developed a physics-informed neural network (PINN) incorporating a two-compartment PK model. The four parameters of the PK model are predicted by a NN in which the numerical representation of the light and heavy chain sequences derived through sequence-attention and protein language model features serve as input. The model successfully predicted clearance and volume of distribution at steady state with remarkable accuracy, achieving predictions within 2-fold error for 10/13 and 13/13 mAbs in the test set, respectively. Concentration-time profiles were well-characterized with a median geometric mean fold error (GMFE) close to 2, with nearly 70% of predictions having GMFE less than 2. This proof-of-concept study demonstrates the feasibility of sequence-based PK prediction for mAbs, potentially reducing animal testing requirements while accelerating candidate selection in therapeutic antibody development. The approach aligns with regulatory initiatives promoting new approach methodologies and complements existing sequence-based predictions of protein structure and target binding during the design phase.