Dose-Response Profiling in Heterologous Gene Expression: Insights from Proteome Fraction Analysis
Dose-Response Profiling in Heterologous Gene Expression: Insights from Proteome Fraction Analysis
Arias Vaccari, N.; Zevallos-Aliaga, D.; Peeters, T.; Guerra Giraldez, D.
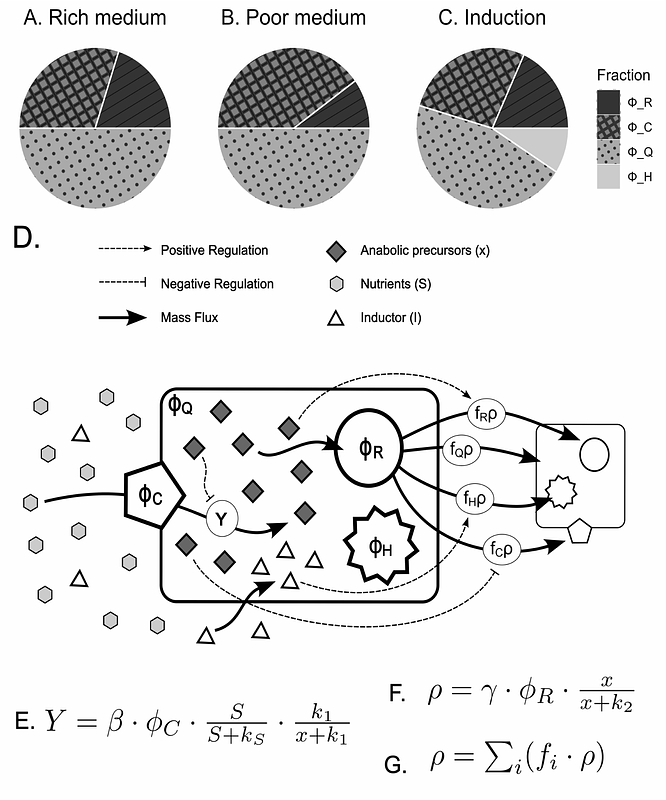
AbstractTo achieve precise control over the expression of heterologous genes, crucial to applications like biosensors and protein manufacturing, it is essential to accurately measure the systems response to varying levels of the inducer. This study introduces a formal approach, inspired by proteome fraction concepts, to analyze titration experiments with a molecular inducer. By deriving the proteome fraction definition with respect to time, we introduce the concept of expression fraction, representing the portion of the anabolic rate dedicated to the production of a specific set of proteins. Formal deduction reveals that the expression fraction equals the proteome fraction when it is at its maximum point, enabling its reliable measurement through direct quantitation of protein amounts. Additionally, experimental data from microcultures and fluorescent signals in three biosensors and six cellular contexts show a linear correlation between protein production and growth rate during exponential growth, indicating a constant expression fraction through this time window. The values of expression fraction obtained from the slopes of such intervals or those obtained from maximum points of protein amount can be fitted independently to a Hill function and produce remarkably similar dose-response parameters. Thus, this conceptual framework provides two consistent methods for determining regulatory parameters. The model is further applied to explore possible mechanisms for how the heterologous expression negatively impacts growth rates in different strains, with Escherichia coli B strains generally exhibiting greater tolerance than K12.