Automated evaluation of single-cell reference atlas mappings enables the identification of disease-associated cell states
Automated evaluation of single-cell reference atlas mappings enables the identification of disease-associated cell states
Sikkema, L.; Lkandushi, M.; Scarcella, D.; Moinfar, A. A.; Engelmann, J. P.; Theis, F. J.
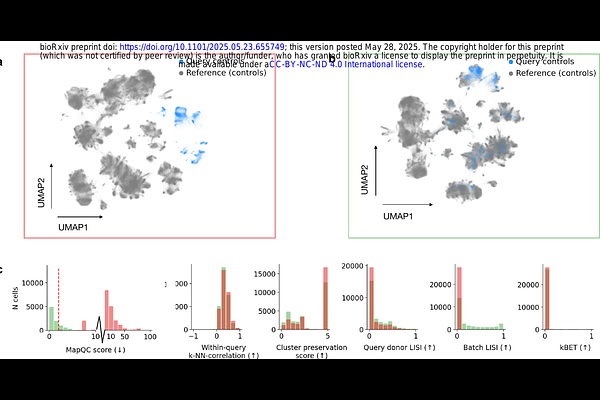
AbstractThe rise of single-cell atlases has opened up the use of reference atlases as a comprehensive healthy control for the study of disease. A key step for using atlases is the mapping of \"query\" disease or perturbed datasets onto a healthy reference atlas, which allows for comparison of the query to healthy controls and thereby the identification of perturbation-specific transcriptional cell states. However, the mapping success, i.e. the extent to which these mappings correctly reflect biological similarities and differences between query and reference, varies with e.g. experimental confounders in the data (dissociation protocol, single-cell assay) or the mapping method used, and cannot be predicted in advance. Moreover, an unsuccessful mapping can result in falsely reporting technical artifacts as disease or perturbation-specific cellular changes and in overlooking true altered states. Here, we present MapQC, a method that quantifies query-to-reference mapping success by leveraging the information present in the reference embedding. Specifically, MapQC tests the presence of remaining batch effect in the embedding after mapping by comparing inter-sample distances in the healthy reference itself to distance of query control samples to the reference. Similarly, it tests whether disease or perturbation-specific variation has been retained during mapping by testing whether query perturbed samples are more distant to the reference than reference samples are from each other. We apply MapQC to lung and endometrial query datasets, including data of idiopathic pulmonary fibrosis and Asherman Syndrome, mapped to large-scale healthy references. We show that MapQC correctly distinguishes successful mappings from unsuccessful ones where data visualization techniques such as UMAP can be deceiving, and that mapQC outperforms integration metrics often re-purposed for mapping quality-control. Moreover, only mappings that MapQC identified as successful result in the correct identification of cell state changes specific to disease. Taken together, MapQC provides a critical quality-control metric for a more reliable, robust, and accurate use of reference atlases to study disease.