Hybrid Population PK-Machine Learning Modeling to Predict Infliximab Pharmacokinetics in Pediatric and Young Adult Patients with Crohn's Disease
Hybrid Population PK-Machine Learning Modeling to Predict Infliximab Pharmacokinetics in Pediatric and Young Adult Patients with Crohn's Disease
Irie, K.; Phillip, M.; Reifenberg, J.; Brendan, B. M.; Noe, J. D.; Jeffrey, H.; Mizuno, T.
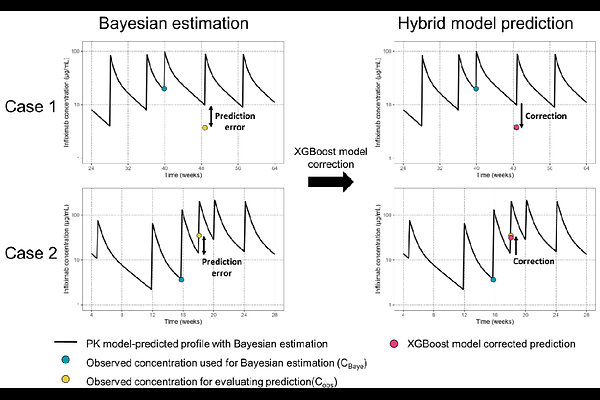
AbstractPopulation pharmacokinetic (PK) model-based Bayesian estimation is widely used for dose individualization, particularly when sample availability is limited. However, its predictive accuracy can be compromised by factors such as misspecified prior information, intra-patient variability, and uncertainties in PK variations. In this study, we developed a hybrid approach that combines machine learning (ML) with population PK-based Bayesian methods to improve the prediction of infliximab concentrations in children with Crohn\'s disease. We calculated prediction errors between Bayesian-estimated and observed infliximab concentrations from 292 measurements across 93 patients. Incorporating clinical patient features, we explored various ML algorithms, including linear regression, random forest, support vector regression, neural networks, and XGBoost to correct the Bayesian-based prediction errors. The predictive performance of these ML models was assessed using root mean square error (RMSE) and mean prediction error (MPE) with 5-fold cross-validation. For Bayesian estimation alone, the RMSE and MPE were 4.8 mcg/mL and -0.67 mcg/mL, respectively. Among the ML algorithms, the XGBoost model demonstrated the best performance, achieving an RMSE of 3.78 +/- 0.85 mcg/mL and an MPE of -0.03 +/- 0.69 mcg/mL in 5-fold cross-validation. The ML-corrected Bayesian estimation significantly reduced the absolute prediction error compared to Bayesian estimation alone. This hybrid population PK-ML approach provides a promising framework for improving the predictive performance of Bayesian estimation, with the potential for continuous learning from new clinical data to enhance dose individualization.