SCLC-TumorMiner: A Directly Accessible Genomics Resource for Precision Oncology: Big Data for Small Cells
SCLC-TumorMiner: A Directly Accessible Genomics Resource for Precision Oncology: Big Data for Small Cells
Elloumi, F.; Dhall, A.; Taniyama, D.; Luna, A.; Yasuhiro, A.; Varma, S.; Wang, J.; Tehim, A.; Raffeld, M.; Aldape, K.; Redon, C.; Shresta, R.; Reinhold, W.; Aladjem, M. I.; Del Rivero, J.; Roper, N.; Thomas, A.; Pommier, Y.
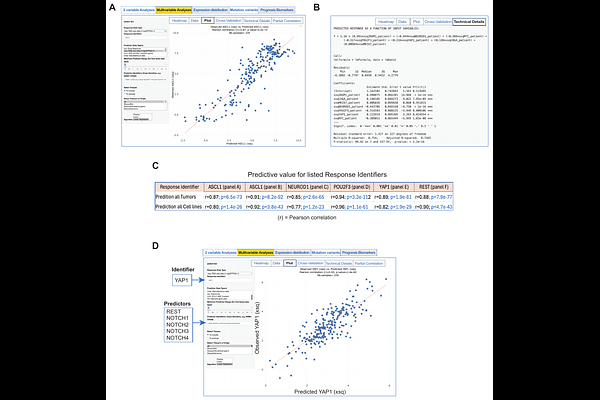
AbstractRNAseq and DNAseq are fast, cost-effective and quantitative methods to dissect cancer cells. However, for each sample, they generate thousands of data points, and comparing patient samples multiplies the complexity. To handle these difficulties, we developed TumorMiner, a web-based tool for clinicians and basic researchers. Here we present our analyses and website for Small Cell Lung Cancer (SCLC). SCLC_TumorMiner includes 235 samples from untreated and relapse patients across the NCI, the University of Rochester, Tongji University and the University of Cologne (https://discover.nci.nih.gov/SclcTumorMinerCDB/). SCLC_TumorMiner allows the molecular classification of tumors based on the canonical NAPY classification and NMF, genomic network analyses exemplified by the Myc and Notch pathways, and the identification of risk-factors and predictive gene expression biomarkers for cell surface and intracellular targets such as DLL3, TROP2, SEZ6, CEA, TRPM5, SLFN11 and proapoptotic and multidrug resistance genes. The architecture of TumorMiner allows its extension to proteomic databases, and to other cancers and institutions worldwide to elucidate cancer pathways, achieve precision medicine and serve as a medical assistant software.