Co-mutation based genetic networks to infer temporal mutation dynamics in ancient human mitochondrial genomes
Co-mutation based genetic networks to infer temporal mutation dynamics in ancient human mitochondrial genomes
Verma, R. K.; Fuentes-Gonzalez, J.; Pienaar, J.; Fierst, J. L.
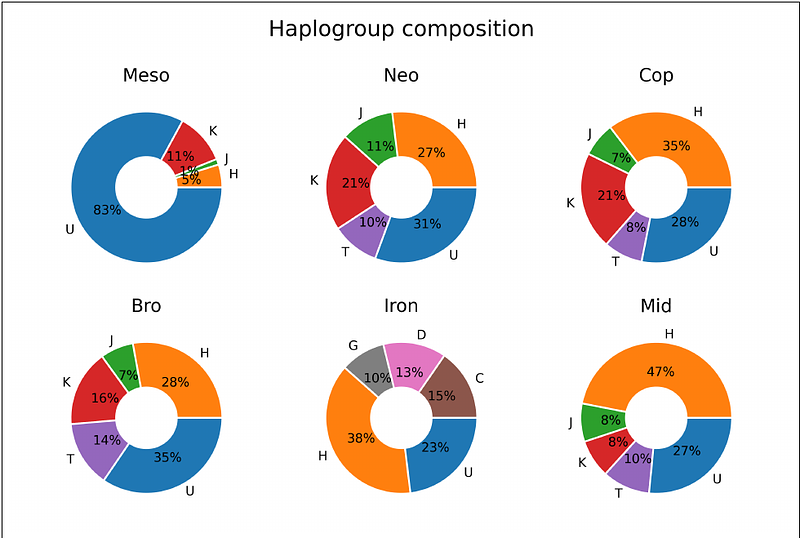
AbstractThe evolution of modern humans is as fascinating as it is complicated. Various environmental and socio-political factors have played a significant role in shaping different migratory paths and lifestyles in ancient times and the establishment of current sub-populations around the globe. These factors have impacted the molecular evolution of mitochondrial DNA (mtDNA). To account for the role of genetic background and variable sites in shaping the migratory and evolutionary paths, we analyzed the ancient mtDNA as spatiotemporal co-mutation networks. Haplogroup-based analysis of the variable sites suggests a transition from hunter-gatherer to agrarian lifestyle during the Copper-Bronze transition period. Furthermore, the genetic interaction networks showed that COX and CYB genes behaved diversely with time and that there is an interplay of the genetic interactions with NADH Dehydrogenase genes specific to each age. We performed a phylogeny (tree) based gene network analysis and examined polymorphism to divergence ratios to complement the co-mutation networks. The tree-based gene networks exhibit similar topology with reduced co-mutation among the genes. The more traditional polymorphism/divergence analysis indicated that the CYB gene has been under long-term purifying selection. In contrast, other genes (ATP6, COX, NADH hydrogenases) have undergone episodes of purifying selection corresponding to different ages. We believe this network-based exploratory study provides insights into early lifestyle transitions and haplogroup establishment of the current human population.