Positional frequency chaos game representation for machine learning-based classification of crop lncRNAs
Positional frequency chaos game representation for machine learning-based classification of crop lncRNAs
Papastathopoulos-Katsaros, A.; Liu, Z.
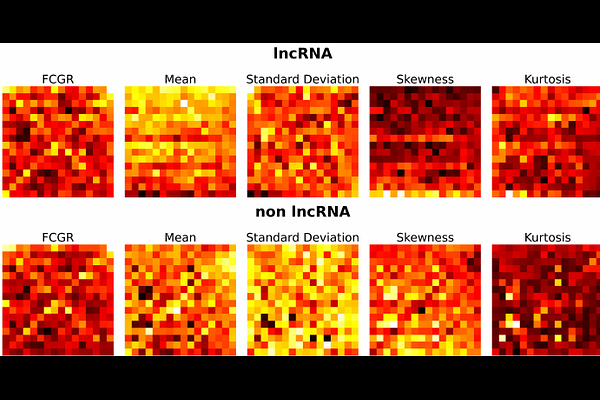
AbstractMotivation: Alignment-based methods are fundamental for sequence comparison but are often computationally prohibitive for large-scale genomic analyses. This limitation has spurred the development of quicker, alignment-free alternatives, such as k-mer analysis, which are crucial for studying long non-coding ribonucleic acids (lncRNAs) in plants. These lncRNAs play critical roles in regulating gene expression at both the epigenetic and transcriptomic levels. However, existing alignment-free approaches typically lose positional information, which can be vital for achieving accurate classification. Results: We propose positional frequency chaos game representation (PFCGR), a novel encoding that improves the traditional frequency chaos game representation (FCGR) by incorporating four statistical moments of k-mer positions: mean, standard deviation, skewness, and kurtosis. This creates a multi-channel image representation of genomic sequences, enabling machine learning models such as Logistic Regression, Random Forests, and Convolutional Neural Networks to classify plant lncRNAs directly from raw genomic sequences. Tested on seven major crop species, our PFCGR-based classifiers achieve classification accuracies comparable to or exceeding those of the computationally intensive DNABERT-based model, while requiring significantly less computational resources. These results demonstrate PFCGR\'s potential as an efficient and accurate tool for plant lncRNA identification, as well as its ability to facilitate large-scale computational studies in genomics.