Parallelization of single-molecule binding kinetic measurements via protein barcode sequencing
Parallelization of single-molecule binding kinetic measurements via protein barcode sequencing
Hutchinson, S.; Mansour, G. H.; Rehan, A.; Pichard-Kostuch, A.; Redheuil, E.; Reed, B. D.; Griffiths, A. D.; Ribezzi-Crivellari, M.
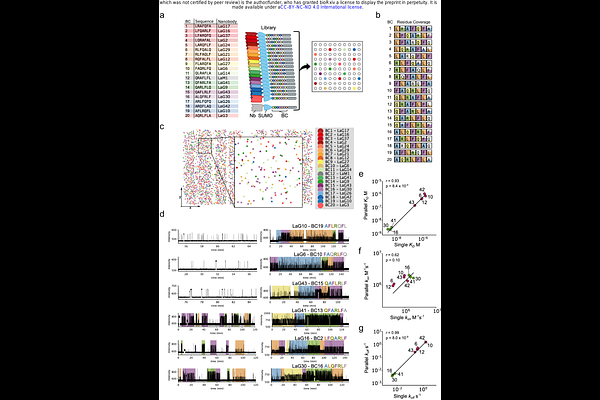
AbstractScreening protein variants for desired functions has long relied on coupling of genotype (gene sequence) to phenotype (protein function), limiting the use of powerful single-molecule (SM) techniques. Here, we introduce a scalable SM screening method that bypasses this constraint by linking SM functional analysis to protein identity through SM protein sequencing. Protein variants are tagged with unique C-terminal peptide barcodes and loaded onto a semiconductor chip containing millions of nanowells. Protein-ligand interactions are monitored in real time at the SM level, and a dye-cycling strategy extends the measurable dynamic range, enabling quantification of slow dissociation rates typical of high-affinity interactions. After functional analysis, each protein molecule is identified by sequencing its barcode. We apply this method to 20 barcoded nanobodies spanning over 1,000-fold in affinity, yielding results consistent with published values and individual SM measurements. Our approach should accelerate protein engineering by enabling rapid, multiplexed SM screening of protein libraries.