Visible-Light-Controlled Residue-Selective Cross-Linking for Deciphering Protein Complexes and Dynamic Protein-Protein Interactions in Live Cells
Visible-Light-Controlled Residue-Selective Cross-Linking for Deciphering Protein Complexes and Dynamic Protein-Protein Interactions in Live Cells
Xu, Y.; Hu, H.; Ran, Y.; Zhao, W.; Guo, A.-D.; Nie, H.-J.; Zhai, L.; Yin, G.-L.; Cheng, J.-T.; Tao, S.; Yang, B.; Tan, M.; Chen, X.-H.
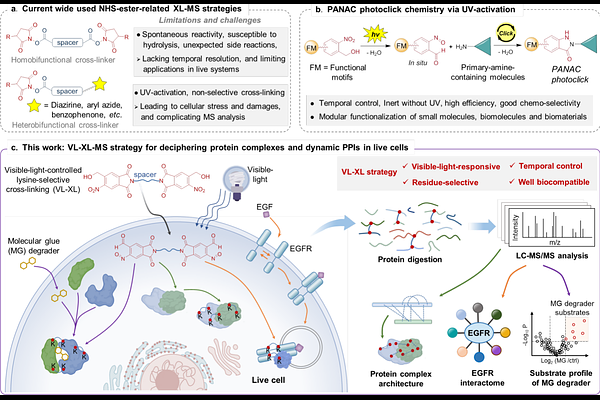
AbstractCross-linking mass spectrometry (XL-MS) has emerged as an attractive technology for investigating protein complexes and protein-protein interactions (PPIs). However, commonly used cross-linking strategies present significant challenges for precise analysis of protein complexes and dynamic PPIs in native biological environments. Here we present the visible-light-controlled lysine-selective cross-linking (VL-XL) strategy for in-depth analysis of protein complexes and dynamic PPIs both in vitro and in live cells, building on light-induced primary amines and o-nitrobenzyl alcohols cyclization (PANAC) chemistry. We demonstrate that the VL-XL strategy effectively explores the dynamic dimerization of PD-L1 stimulated by exogenous modulators. Moreover, the VL-XL strategy successfully profiles the time-resolved EGF-stimulated EGFR interactome, providing valuable insights into the regulatory mechanisms of EGFR signaling and intracellular trafficking. Importantly, the VL-XL strategy efficiently deciphers the molecular glue (MG) induced dynamic PPIs and substrate profile of MG degrader, opening an innovative avenue for identifying neo-substrates. By harnessing the advantages of temporal controllability, good biocompatibility, and lysine selectivity, the VL-XL method simplifies MS data analysis and facilitates the acquisition of accurate structural information of protein complexes and the elucidation of elusive PPIs in live cells. Overall, the VL-XL strategy expands the XL-MS toolbox, and realizes in-depth analysis of protein complexes and dynamic PPIs, which will inspire innovative solutions for protein interactomes research and structural systems biology.