Iterative dispersion update achieves global optimum fitting linear mixed model in genetic associations
Iterative dispersion update achieves global optimum fitting linear mixed model in genetic associations
Guan, Y.; Levy, D.
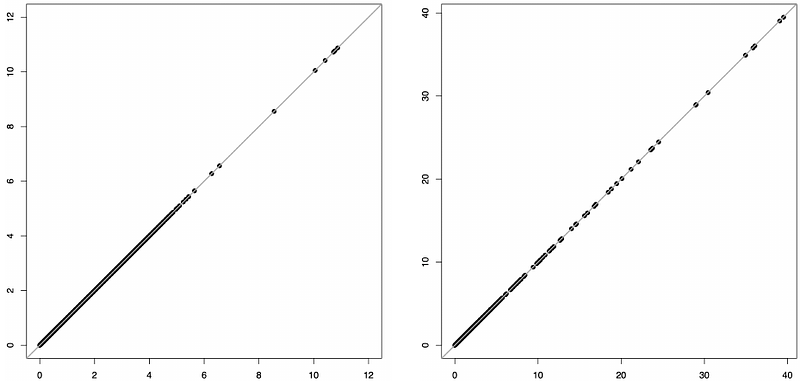
AbstractIn genetic association studies, the linear mixed model (LMM) has become a standard practice to account for population stratification and relatedness in the samples to reduce false positives. Despite recent progress, fitting LMMs to achieve an exact optimum has no theoretical assurance and remains computationally demanding, particularly in multi-omics studies where tens of thousands of phenotypes are tested for association with millions of genetic markers. The state-of-the-art method used the Newton-Raphson algorithm for optimization. Here we present IDUL that uses iterative dispersion updates to fit LMMs. IDUL requires no evaluation of derivatives of the likelihood function, rather, it fits two weighted least square regressions in each iteration. Applied to a set of protein assays from the Framingham Heart Study, IDUL converged in several iterations and provided consistent estimates between different initial values, which outperformed the Newton-Raphson method. IDUL is highly efficient for additional phenotypes, making it ideal to study genetic determinant of multi-omics phenotypes. Through a theoretical lens, we show that IDUL is a gradient ascent algorithm, and is as efficient as the Newton-Raphson method near the optimum. Most significantly, with a sufficiently large sample, IDUL converges to the global optimum with probability one, achieving the exact optimum. IDUL is freely available at http://www.haplotype.org.