FAIMS-GPF XL-MS: crosslinking-mass spectrometry based on gas-phase fractionation
FAIMS-GPF XL-MS: crosslinking-mass spectrometry based on gas-phase fractionation
Gizardin-Fredon, H.; Terrosu, S.; Pichard, S.; Korkut, D.; Braun, C.; Poterszman, A.; Charenton, C.; Cianferani, S.
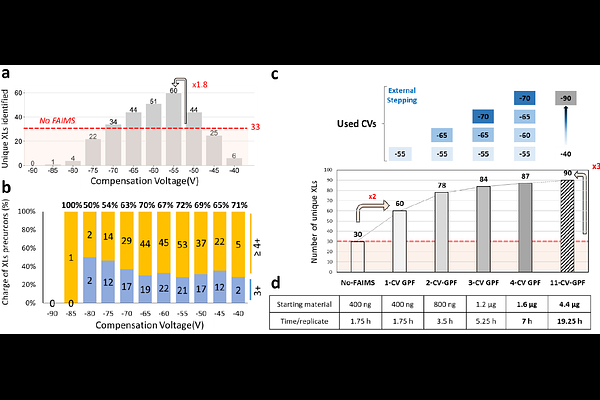
AbstractProtein-protein interactions (PPIs) underpin nearly all cellular processes; therefore, mapping PPI and protein-protein association networks is critical for understanding how biological systems function in health and disease. However, many functionally relevant PPIs are transient and mediated by weak affinity interactions, making them challenging to detect. Using chemical cross-linking together with mass spectrometry (XL-MS) has emerged as an important category of methods for mapping PPIs, including those that are more dynamic and transient, although XL-MS workflows face limitations which hinder their routine use, especially in proteomic platforms. Here, to address these limitations, we developed a XL-MS workflow based on gas-phase fractionation (GPF) using high-field asymmetric waveform ion mobility spectrometry (FAIMS). Our optimized FAIMS-GPF XL-MS protocols exhibited improved performance when handling both low-complexity samples, such as purified Cdk7-Activating Kinase (CAK) heterotrimer, and high-complexity cross-linked HeLa lysates. In HeLa lysate, we identified 1,278 cross-links accounting for 213 PPIs without any in-solution fractionation, starting from less than 20 ug of sample. The method also proved effective for analyzing low-abundance, high-complexity samples such as spliceosomes. Overall, FAIMS-GPF enhanced the detection of cross-linked peptides and improved PPI coverage, thus offering a scalable, high-sensitivity solution for XL-MS integration with structural biology and functional proteomics applications.