LightRoseTTA: High-efficient and Accurate Protein Structure Prediction Using an Ultra-Lightweight Deep Graph Model
LightRoseTTA: High-efficient and Accurate Protein Structure Prediction Using an Ultra-Lightweight Deep Graph Model
Wang, X.; Zhang, T.; Liu, G.; Cui, Z.; Zeng, Z.; Long, C.; Zheng, W.; Yang, J.
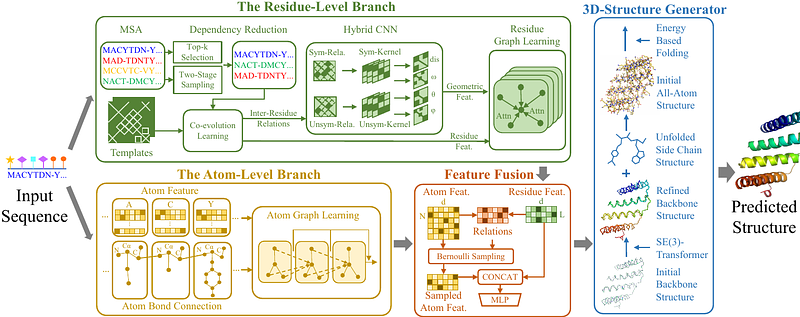
AbstractAccurately predicting protein structure, from amino acid sequences to three-dimensional structures, is of great significance in biological research. To tackle this issue, a representative deep big model, RoseTTAFold, has been proposed with promising success. Here, we report an ultra-lightweight deep graph network, named LightRoseTTA, to achieve accurate and high-efficient prediction for proteins. Notably, three highlights are possessed by our LightRoseTTA: (i) high-accurate structure prediction for proteins, being competitive with RoseTTAFold on multiple popular datasets including CASP14 and CAMEO; (ii) high-efficient training and inference with an ultra-lightweight model, costing only one week on one single general NVIDIA 3090 GPU for model-training (vs 30 days on 8 high-speed NVIDIA V100 GPUs for RoseTTAFold) and containing only 1.4M parameters (vs 130M in RoseTTAFold); (iii) low dependency on multi-sequence alignments (MSA, widely-used homologous information), achieving the best performance on three MSA-insufficient datasets: Orphan, De novo, and Orphan25. Besides, our LightRoseTTA is transferable from general proteins to antibody data, as verified in our experiments. We visualize some case studies to demonstrate the high-quality prediction, and provide some insights on how the structure predictions facilitate the understanding of biological functions. We further make a discussion on the time and resource costs of LightRoseTTA and RoseTTAFold, and demonstrate the feasibility of lightweight models for protein structure prediction, which may be crucial in the resource-limited research for universities and academy institutions. We release our code and model to speed biological research.