Single cell multiomic analysis of the impact of Delta-9-tetrahydrocannabinol on HIV infected CD4 T cells
Single cell multiomic analysis of the impact of Delta-9-tetrahydrocannabinol on HIV infected CD4 T cells
Ashokkumar, M.; Ge, R. Y.; Cooper-Volkheimer, A.; Margolis, D.; Li, Q.; Jiang, Y.; Murdoch, D. M.; Browne, E. P.
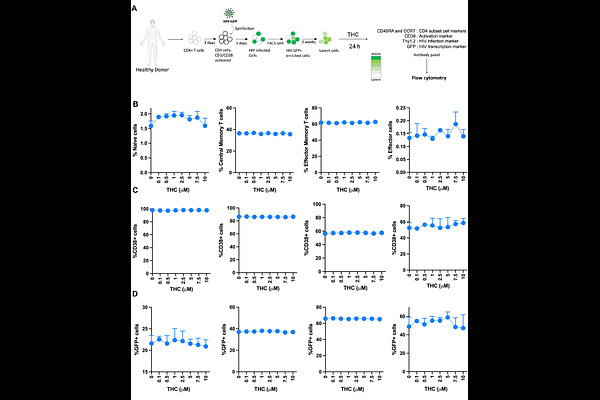
AbstractCannabis use is prevalent among individuals living with HIV in the United States, but the impact of cannabis exposure on the reservoir of latently infected cells that persists during antiretroviral therapy (ART) remains unclear. To address this gap, we analyzed the effect of D-9-tetrahydrocannabinol (THC) on primary CD4 T cells that were latently infected with HIV. We found that THC had no detectable effect on baseline or latency reversing agent (LRA) stimulated HIV expression, or on expression of an activation marker (CD38). However, using an integrated multiomic single-cell analysis of genome-wide chromatin accessibility and gene expression, we observed altered expression of several hundred genes in HIV infected CD4 T cells after THC exposure, including transcriptional downregulation of genes involved in protein translation and antiviral pathways, indicating that THC suppresses innate immune activation in infected cells. Additionally, chromatin accessibility analysis demonstrated upregulated chromatin binding activity for the transcriptional regulator CTCF, and reduced activity for members of the ETS transcription factor family in infected cells after THC exposure. These findings provide insights into the mechanisms by which cannabis use could influence the persistence of HIV within cellular reservoirs and the molecular phenotype of latently infected cells. Further elucidation of the underlying mechanisms involved in THC-mediated changes to HIV infected cells, will lead to an improved understanding of the impact of cannabis use on the HIV reservoir.