GREmLN: A Cellular Regulatory Network-Aware Transcriptomics Foundation Model
GREmLN: A Cellular Regulatory Network-Aware Transcriptomics Foundation Model
Zhang, M.; Swamy, V.; Cassius, R.; Dupire, L.; Kanatsoulis, C.; Paull, E.; AlQuraishi, M.; Karaletsos, T.; Califano, A.
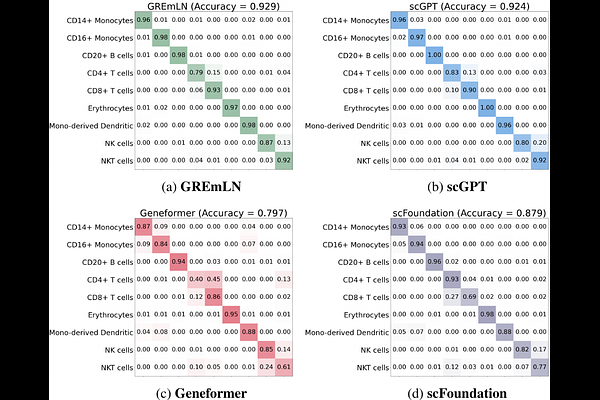
AbstractThe ever-increasing availability of large-scale single-cell profiles presents an opportunity to develop foundation models to capture cell properties and behavior. However, standard language models such as transformers are best suited for sequentially structured data with well defined absolute or relative positional relationships, while single cell RNA data have orderless gene features. Molecular-interaction graphs, such as gene regulatory networks (GRN) or protein-protein interaction (PPI) networks, offer graph structure-based models that effectively encode both non-local gene-gene dependencies, as well as potential causal relationships. We introduce GREmLN, a foundation model that leverages graph signal processing to embed gene-regulatory network structure directly within its attention mechanism, producing biologically informed single cell specific gene embeddings. Our model faithfully captures transcriptomics landscapes and achieves superior performance relative to state-of-the-art baselines on both cell type annotation and graph structure understanding tasks. It offers a unified and interpretable framework for learning high-capacity foundational representations that capture complex, long-range regulatory dependencies from high-dimensional single-cell transcriptomic data. Moreover, the incorporation of graph-structured inductive biases enables more parameter-efficient architectures and accelerates training convergence.