The epigenetics effects of transposable elements are context dependent and not restricted to gene silencing
The epigenetics effects of transposable elements are context dependent and not restricted to gene silencing
Coronado-Zamora, M.; Gonzalez, J.
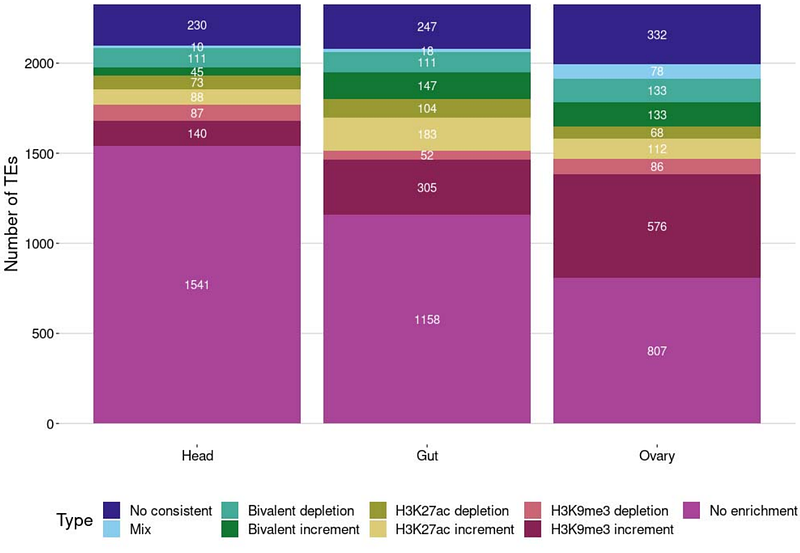
AbstractTransposable elements (TEs) represent a threat to genome integrity due to their proliferation capacity. Eukaryotic cells silence TEs through different epigenetic mechanisms, including the deposition of repressive histone marks. Previous studies have shown that repressive marks can spread to neighboring sequences. However, evidence for this spreading affecting nearby gene expression remains limited. Similarly, whether TEs induce changes in the enrichment of active histone marks genome-wide, and its potential impact on gene expression have not been widely studied. In this work, we performed a comprehensive study of the epigenetic effects of 2,235 TEs and their potential effects on nearby gene expression on D. melanogaster head, gut and ovary. While most of the TEs (816) induce the enrichment of the H3K9me3 repressive mark, with stronger epigenetic effects in the ovary, a substantial number (345 TEs) induce the enrichment of the H3K27ac active mark, particularly in the gut. We found that 70% of the H3K9me3 enriched TEs induced gene down-regulation, and 50% of the H3K27ac enriched TEs induced gene up-regulation. These changes in expression affect specific regulatory networks in head and gut while in ovary, genes were not enriched for any biological functions. Furthermore, TE epigenetic effects on gene expression are genomic context dependent. Finally, we found that TEs also affect gene expression by disrupting regions enriched for histone marks. Overall, our results show that TEs do generate regulatory novelty through epigenetic changes, with these epigenetic effects not restricted to gene silencing and being context dependent.