Highly Sequence-specific, Timing-controllable m6A Demethylation by Modulating RNA-binding Affinity of m6A Erasers
Highly Sequence-specific, Timing-controllable m6A Demethylation by Modulating RNA-binding Affinity of m6A Erasers
Otonari, K.; Asami, Y.; Ogata, K.; Ishihama, Y.; Futaki, S.; Imanishi, M.
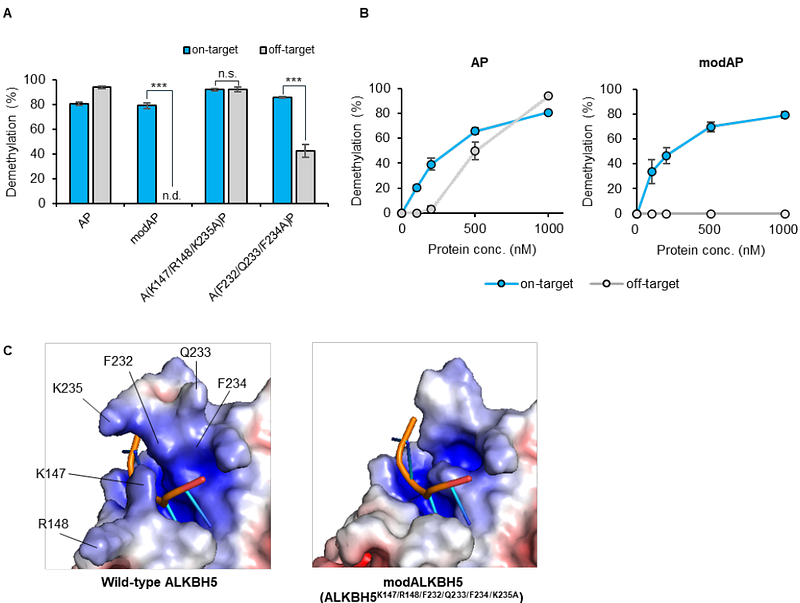
AbstractIn recent years, significant progress has been made in developing tools that consist of programmable RNA binding proteins and m6A-erasers to selectively demethylate m6A at specific sites. Especially, timing-controllable demethylation tools using stimulus-inducible dimerization show promise in understanding the impact of individual m6A modifications on dynamic physiological processes. However, a critical issue with these technologies is the possible off-target effects, wherein wild-type m6A-erasers constituting the tools may decrease methylation levels at sites other than the intended target m6A. In this study, we addressed this issue by reducing the intrinsic RNA-binding ability of m6A-erasers to prevent off-target effects and by fusing with PUF RNA-binding protein to provide recognition ability of arbitrary RNA sequences. Taking advantage of the reduced RNA-binding ability of the m6A-erasers themselves, we employed a rapamycin-based switching system in the linker connecting the PUF and m6A-eraser domains. The resulting m6A-erasers exhibited superior precision in demethylating targeted sites while minimizing off-target effects, presenting a novel approach for precise temporal control over m6A modification dynamics.


