Increasing certainty in systems biology models using Bayesian multimodel inference
Increasing certainty in systems biology models using Bayesian multimodel inference
Linden-Santangeli, N.; Zhang, J.; Kramer, B.; Rangamani, P.
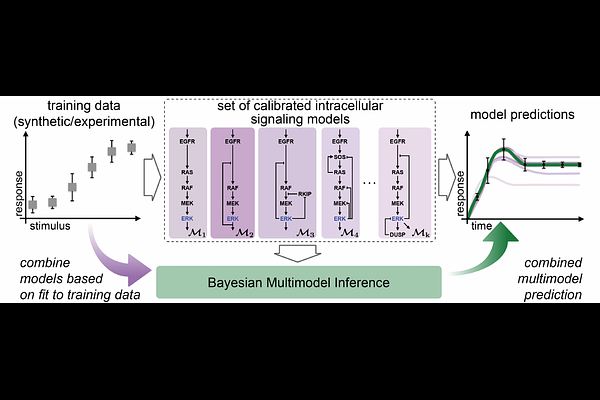
AbstractMathematical models are indispensable to the system biology toolkit for studying the structure and behavior of intracellular signaling networks. A common approach to modeling is to develop a system of equations that encode the known biology using approximations and simplifying assumptions. As a result, the same signaling pathway can be represented by multiple models, each with its set of underlying assumptions, which opens up challenges for model selection and decreases certainty in model predictions. Here, we use Bayesian multimodel inference to develop a framework to increase certainty in systems biology models. Using models of the extracellular regulated kinase (ERK) pathway, we first show that multimodel inference increases predictive certainty and yields predictors that are robust to changes in the set of available models. We then show that predictions made with multimodel inference are robust to data uncertainties introduced by decreasing the measurement duration and reducing the sample size. Finally, we use multimodel inference to identify a new model to explain experimentally measured sub-cellular location-specific ERK activity dynamics. In summary, our framework highlights multimodel inference as a disciplined approach to increasing the certainty of intracellular signaling activity predictions.