Hierarchical prediction and perturbation of chromatin organization reveal how loop domains mediate higher-order architectures
Hierarchical prediction and perturbation of chromatin organization reveal how loop domains mediate higher-order architectures
Wei, J.; Xue, Y.; Gao, Y. Q.
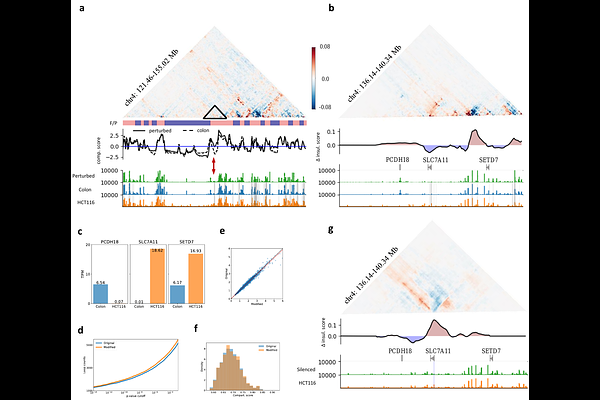
AbstractThe genome is folded within the dense cell nucleus in a hierarchical manner, resulting in complex interactions between distinct folding strategies at various length scales. To elucidate how short-range loop domains regulate higher-order structures of the chromatin, such as topologically associating domains (TADs) and compartments, we introduce HiCGen, a hierarchical and cell-type-specific generator based on Swin-transformer architecture. HiCGen predicts genome organization across different spatial scales utilizing DNA sequence and genomic features as inputs. The model enables in silico screening through genetic or epigenetic perturbations on genome architecture, with resolution down to 1 kb. Our analysis reveals unexpected linear correlations between loop properties and genome organization at various levels, including insulation degree, compartmentalization, and contact intensity over genomic distances exceeding 10 Mb. Regional or global perturbation conducted by HiCGen provides biological implications for such cross-scale correlations and their genome-function dependence. Notably, perturbation analysis of the human genome in sigmoid colon tissue demonstrates that modest activation of carcinogenesis-associated enhancers is sufficient to hijack nearby promoter, reshape TAD boundaries, and even flip compartment at mega-base scale.