Pangebin: improving plasmid binning in bacterial isolates using pangenome-assembly graphs.
Pangebin: improving plasmid binning in bacterial isolates using pangenome-assembly graphs.
Sgro, M.; Brejova, B.; Vinar, T.; Priola, Y.; Bonizzoni, P.; Chauve, C.
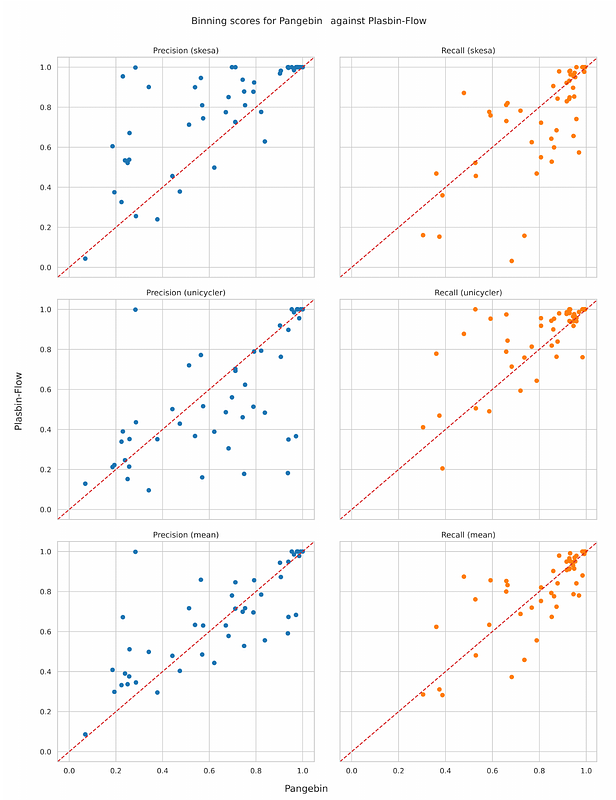
AbstractShort-read genome assemblies typically consist of many contigs of variable lengths and their putative connections represented as an assembly graph. Assembly graphs produced by different tools from the same data may differ significantly, posing a challenge to tools for downstream processing tasks. One such task is plasmid binning, that is identifying plasmids in sequenced bacterial isolates, which is crucial for monitoring the spread of antimicrobial resistance. When plasmid binning tools are applied to assembly graphs produced by different tools, they may exhibit different performance, and choosing the best results a priori can be difficult. To address the above issue, we propose the use of a pangenome graph, built from assembly graphs produced by assembling short reads of the same sample with different assemblers. The resulting pangenome-assembly graph highlights similarities between contigs from different assemblies while retaining information on contigs that appear only in one of the input assemblies. We then used the PlasBin-flow plasmid binning tool customized to take into account pangenome information to identify plasmid bins. The results for pangenome-assemblies built by Unicycler and Skesa show an increase in accuracy measures compared to the mean results obtained on single assemblies, leading to an overall more accurate prediction than a blind choice of assemblers. The source code of the pipeline is available at https://github.com/AlgoLab/pangebin along with the dataset used in this study.