mzrtsim: Raw Data Simulation for Reproducible Gas/Liquid Chromatography Mass Spectrometry Based Non-targeted Metabolomics Data Analysis
mzrtsim: Raw Data Simulation for Reproducible Gas/Liquid Chromatography Mass Spectrometry Based Non-targeted Metabolomics Data Analysis
Yu, M.; Philip, V.
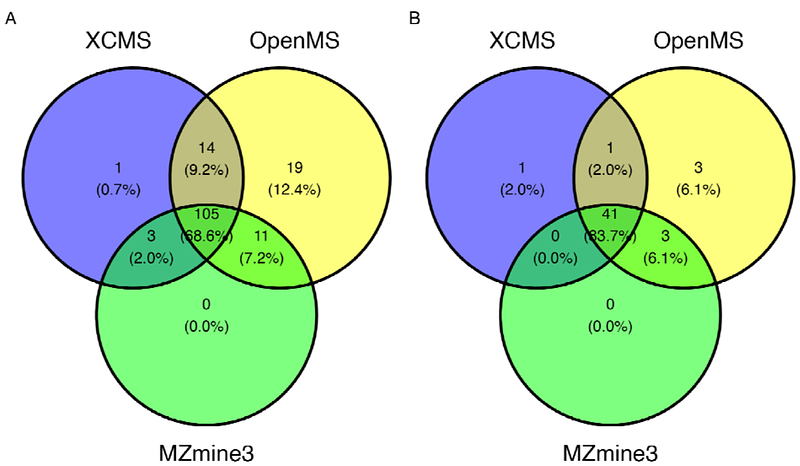
AbstractReproducibility of data analysis is pivotal in the context of non-targeted metabolomics based on mass spectrometry. While various algorithms have been proposed for feature extraction, their validation often revolves around a limited set of known compounds or standards. While data simulation is widely used in other omics studies, many simulations are focused on the feature level, neglecting uncertainties inherent in the feature extraction process for metabolomics mass spectrometry data. In this study, we introduce a R package called \'mzrtsim\' to simulate MS1 full scan raw data in the mzML format. Unlike simulations solely based on statistical distributions, our approach leverages experimental data from the MassBank of North America(MoNA) and the human metabolome database(HMDB). We also developed algorithms to realistically simulate chromatography peaks, accounting for factors such as tailing and leading. The results of our study demonstrate the potential of this tool not only for comparing established metabolomics software (e.g., XCMS, mzMine, and OpenMS) against a ground truth, but also for evaluating their performance under challenging chromatographic conditions often encountered in non-targeted metabolomics. We found that the investigated software all introduced large amounts of false positive peaks (more than 75% of detected peaks by default setting) and loss compounds with less peaks. They also showed different sensitivity to trailing and leading peaks and we also found setting appropriate intensity cutoffs will reduce the influences from redundant peaks. This R package is free and available online(https://github.com/yufree/mzrtsim).