From Atoms to Fragments: A Coarse Representation for Functional and Efficient Protein Design
From Atoms to Fragments: A Coarse Representation for Functional and Efficient Protein Design
Castorina, L. V.; Wood, C. W.; Subr, K.
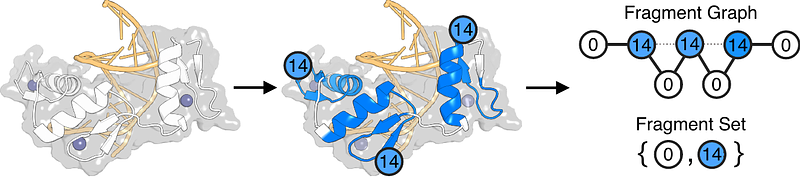
AbstractDeep learning has made remarkable progress in protein design, yet current protein representations remain largely black-box and scale poorly with protein length, leading to high computational costs. We propose a fragment-based protein representation that balances interpretability and efficiency. Using a curated set of 40 evolutionarily conserved fragments, we represent proteins as fragment sets or fragment graphs, significantly reducing dimensionality while preserving functional information. Here, we show that fragment-based representations capture significantly more information at much lower dimensions compared to traditional methods. On a dataset of 215 functionally diverse proteins, our approach outperforms traditional sequence- and structure-based methods in clustering by protein function at <=30% sequence identity. Additionally, fragment-based search achieves comparable accuracy while using 90% fewer tokens. It also runs ~68.7x faster than RMSD-based methods and ~1.64x faster than sequence-based methods, even when including fragment pre-processing overhead. Finally, we show that fragments can guide RFDiffusion backbone generation, with recovery rates higher than 40%. We propose fragment-based representations as a scalable and interpretable alternative for the next generation of protein design tools, spanning backbone and sequence design to functional searches in protein structure databases.