Genome sequencing confirms cryptic diversity and potentially adaptive changes in gene content of Lepidoptera-infecting Apicomplexa
Genome sequencing confirms cryptic diversity and potentially adaptive changes in gene content of Lepidoptera-infecting Apicomplexa
Mongue, A. J.; Kuperus, P.; Groot, A. T.
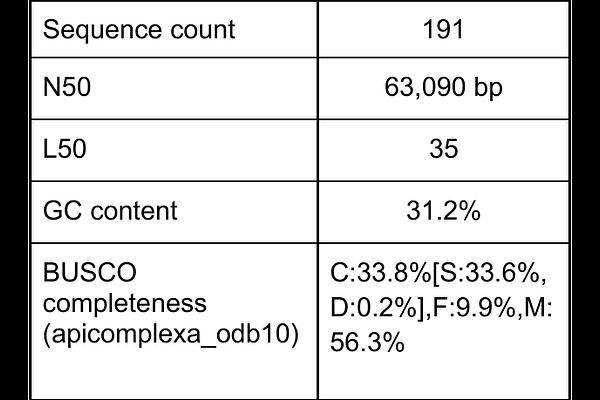
AbstractApicomplexan parasites of insects represent a largely unexplored branch of the tree of life. Their small size and lack of informative phenotypic traits can obscure their diversity and many lineage-specific adaptations to different host species. To better understand how these parasites have evolved, we have sequenced and annotated the genome of a neogregarine parasite of moths including Helicoverpa armigera. This parasite, which we term Helicoverpa-infecting Ophryocystis, has a tiny 7 Megabase genome with only 2,200 protein-coding genes. Using phylogenomic methods, we place it as the closest currently sequenced relative to Ophryocystis elektroscirrha and its sister lineage, both of which infect milkweed butterflies. We then explore evolution of the ATPase gene family, which is known to be reduced in other Ophryocystis lineages. Helicoverpa-infecting Ophryocystis has more ATPase genes than other Ophryocystis, which may relate to the interactions of parasites with phytochemistry encountered in their insect hosts. These results demonstrate both the amount of uncertainty remaining in the insect-infecting Apicomplexa as well as the utility of applying modern genomic approaches to their study.