Positive Selection on Rare Variants Underlying the Cold Adaptation of Wild Boar
Positive Selection on Rare Variants Underlying the Cold Adaptation of Wild Boar
Chen, J.; Jakovlic, I.; Sablin, M.; Xia, S.; Xu, z.; Guo, Y.; Kuang, R.; Zhong, J.; Jia, Y.; Thi, T. N. T.; Yang, H.; Ma, H.; Sprem, N.; Liu, D.; Zhao, Y.; Zhao, S.
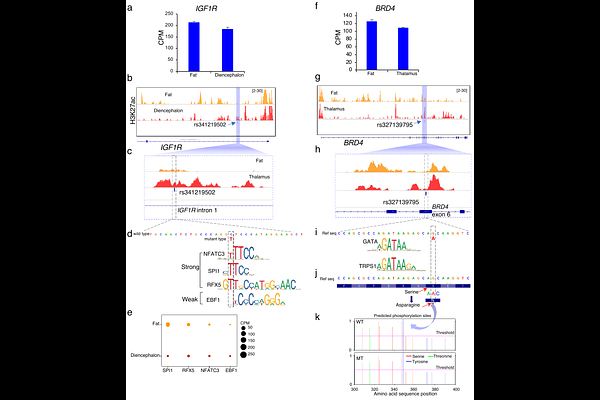
AbstractThe wide geographical distribution of Eurasian wild boar (Sus scrofa) offers a natural experiment to study the thermoregulation. Here, we conducted whole-genome resequencing and chromatin profiling experiments on the local populations from cold regions (northern and northeastern Asia) and warm regions (southeastern Asia and southern China). Using genome-wide scans of four methods, we detected candidate genes underlying cold-adaptation with significant enrichment of pathways related to thermogenesis, fat cell development, and adipose tissue regulation. We also found two enhancer variants under positive selection, an intronic variant of IGF1R (rs341219502) and an exonic variant of BRD4 (rs327139795), which showed the highest differentiation between cold and warm region populations of wild boar and domestic pigs. Moreover, these rare variants were absent in outgroup species and warm-region wild boar but nearly fixed in cold-region populations, suggesting their de novo origins in cold-region populations. The experiments of CUT&Tag chromatin profiling showed that rs341219502 of IGF1R is associated with the gain of three novel transcription factors involving regulatory changes in enhancer function, while rs327139795 of BRD4 could result in the loss of a phosphorylation site due to amino acid alteration. We also found three genes (SLCO1C1, PDE3A, and TTC28) with selection signals in both wild boar and native human populations from Siberia, which suggests convergent molecular adaptation in mammals. Our study shows the adaptive evolution of genomic molecules underlying the remarkable environmental flexibility of wild boar.