Disentanglement of batch effects and biological signals across conditions in the single-cell transcriptome
Disentanglement of batch effects and biological signals across conditions in the single-cell transcriptome
Sakaguchi, S.; Tsutsumi, M.; Nishi, K.; Naoki, H.
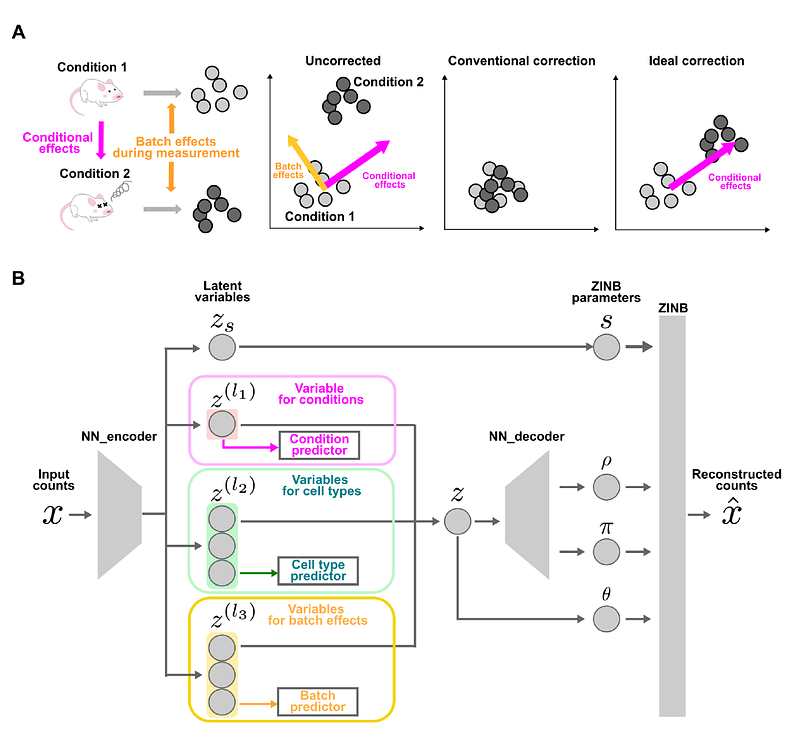
AbstractBatch effects, which arise due to technical variations across different experimental factors, pose a significant challenge for single-cell transcriptomic analysis. Although various batch correction methods have been developed to mitigate these effects, they often indiscriminately mix data from different batches, leading to the removal of biologically meaningful signals. This limitation hinders comparative analyses across multiple conditions, an essential aspect of scientific research. Recent approaches attempt to address this issue by mapping data to separate spaces, but they prevent direct comparisons between conditions. Here, we propose Kanade, a batch correction method based on a variational autoencoder. Kanade explicitly disentangles batch effects from biological signals by specializing latent variables for different types of information. Using both simulated and real datasets, we demonstrate that Kanade selectively correct batch effects while preserving essential biological features, enabling more accurate comparative analyses in single-cell transcriptomics.