Combinatorial Design Testing in Genomes with POLAR-seq
Combinatorial Design Testing in Genomes with POLAR-seq
Ciurkot, K.; Lu, X.; Malyshava, A.; Soro, L.; Lees, A.; Gorochowski, T. E.; Ellis, T.
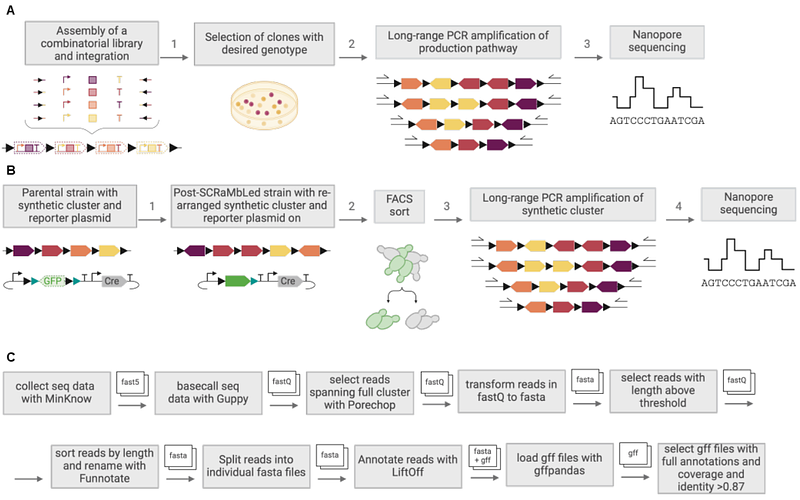
AbstractSynthetic biology projects increasingly use modular DNA assembly or synthetic in vivo recombination to generate diverse combinatorial libraries of genetic constructs for testing. But as these designs expand to multigene systems it becomes challenging to sequence these in a cost-effective way that reveals the genotype to phenotype relationships in the libraries. Here, we introduce a new quick, low-cost method designed for assessing combinational designs of genome-integrated multigene constructs that we call Pool of Long Amplified Reads (POLAR) sequencing. POLAR-seq takes genomic DNA isolated from library pools and uses long range PCR to amplify target genomic regions up to 35 kb long containing combinatorial designs. The pool of long amplicons is then directly read by nanopore sequencing with full length reads then used to identify the gene content and structural variation of individual genotypes in the library and read count indicating how abundant a genotype is within the pool. Using yeast cells with loxP-containing synthetic gene clusters that rearrange in vivo in the presence of Cre recombinase, we demonstrate how POLAR-seq can be used to identify global patterns from combinatorial experiments, find the most abundant genotypes in a pool and also be adapted to sequence-verify gene clusters from isolated strains.


