AlphaFold modeling of polyubiquitin complexes and covalently linked proteins
AlphaFold modeling of polyubiquitin complexes and covalently linked proteins
Fabian, B.; Stuke, J. F. M.; Heinz, M.; Hummer, G.
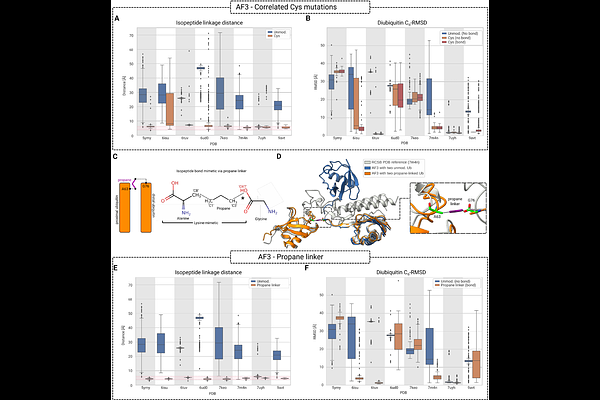
AbstractCells use the covalent attachment of Ubiquitin (Ub) chains to mark proteins for degradation, alter their cellular localization or drive their association. Protein fate is encoded in the distinct poly-Ub linkages, exploiting the vast combinatorial space of linear and branched Ub modifications. AlphaFold has emerged as a powerful tool to predict the structure of protein-protein complexes. However, standard AlphaFold does not consider linkages between individual protein chains, limiting its applicability to Ub chains. The near complete conservation of the ubiquitin sequence and the large number of binding partners suppresses coevolutionary signals, further challenging the prediction of poly-Ub complex structures. We address this challenge, first, by introducing correlated cysteine mutations to induce linkage-specific proximity of Ubs in complex with interacting proteins. Second, we introduce short covalent linker groups in AlphaFold 3 calculations that mimic the isopeptide bonds between linked lysines and Ub C-terminal carboxylates. These two approaches enable the robust structural modeling of complexes involving poly-Ub chains with AlphaFold. The linker approach is general and can be used for other covalent inter-chain connections and to enforce distance restraints for integrative structural modeling.