BaGPipe: an automated, reproducible, and flexible pipeline for bacterial genome-wide association studies
BaGPipe: an automated, reproducible, and flexible pipeline for bacterial genome-wide association studies
Wei, K.; Blane, B.; Toussaint, J.; Reuter, S.; Toleman, M. S.; Torok, E.; Peacock, S. J.; Harrison, E. M.; Aggarwal, D.; Roberts-Sengier, W.
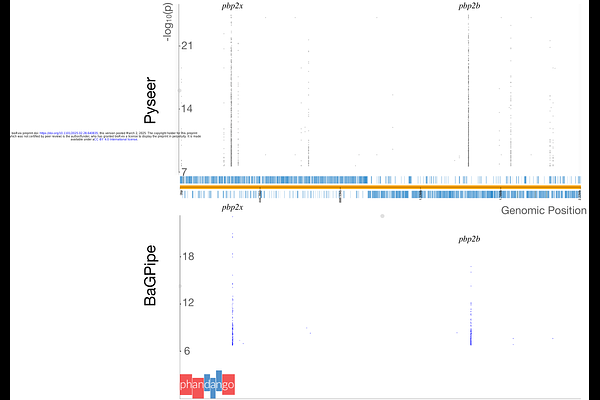
AbstractMicrobial genome-wide association study (GWAS) tools often require manual data processing steps, lack comprehensive workflows, and are limited by scalability issues, thus hindering the exploration of bacterial genetic traits. To address these challenges, we developed BaGPipe, an automated and flexible bacterial GWAS pipeline built using Nextflow and incorporating Pyseer for association analysis. BaGPipe integrates all essential components of a bacterial GWAS--spanning pre-processing, statistical analysis, and downstream visualisation--into a unified workflow that is reproducible and easy to deploy across diverse computational environments. BaGPipe was validated on a publicly available dataset of Streptococcus pneumoniae whole-genome sequences, and reproduced published findings with improved computational efficiency. BaGPipe was then applied to a dataset of Staphylococcus aureus whole-genome sequences, successfully identifying known and novel antibiotic resistance associations. By offering an accessible, efficient, and reproducible platform, BaGPipe accelerates bacterial GWAS and facilitates deeper exploration into the genetic underpinnings of phenotypic traits.