SpaGRN: investigating spatially informed regulatory paths for spatially resolved transcriptomics data
SpaGRN: investigating spatially informed regulatory paths for spatially resolved transcriptomics data
Li, Y.; Liu, X.; Guo, L.; Han, K.; Fang, S.; Wan, X.; Xu, X.; Fan, G.; Xu, M.
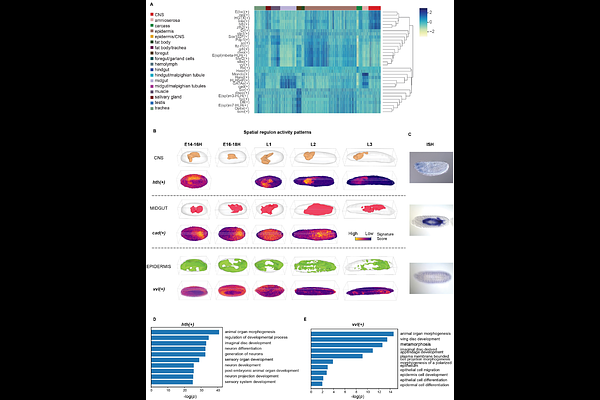
AbstractCells expressing similar transcriptional regulatory circuits spatially aggregate into distinct cell types or states. However, most existing methods for inferring gene regulatory networks from spatially resolved transcriptomics are devoted to spatial co-expression modules or interactions between transcription factors and target genes, neglecting mediated effects from extracellular signals. Here we introduce SpaGRN, a statistical framework for predicting the comprehensive intracellular regulatory network underlying spatial patterns by integrating spatial expression profiles with prior knowledge on regulatory relationships and signaling paths. We validate and assess SpaGRN using simulated and real datasets, demonstrating its efficiency, performance, and robustness. When applied to 3D datasets of developing Drosophila embryos and larvae, SpaGRN identifies spatiotemporal variations in specific regulatory patterns, delineating the cascade of events from receptor stimulation to downstream transcription factors and targets, revealing synergetic regulation mechanism during organogenesis. Moreover, SpaGRN provides flexible visualization functions. We construct an online 3D regulatory network atlas database for interactive exploration and sharing.