Pan-cancer transcriptional regulatory network analysis reveals key drivers and epigenetic modulators in tumorigenesis
Pan-cancer transcriptional regulatory network analysis reveals key drivers and epigenetic modulators in tumorigenesis
Yin, Y.; Zhang, J.
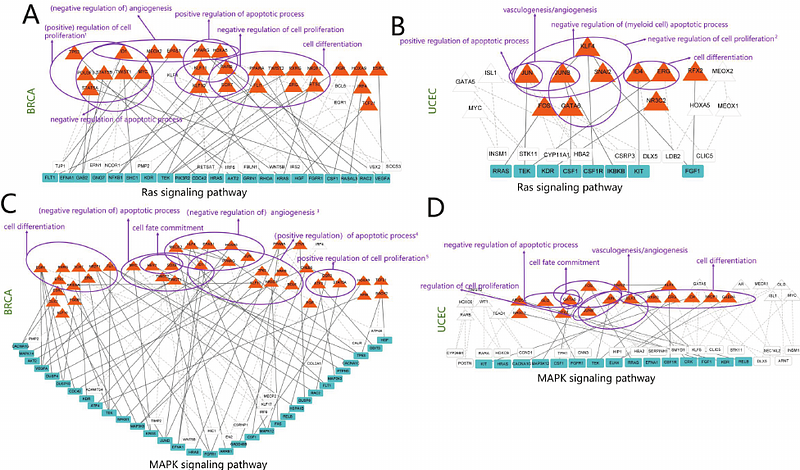
AbstractTranscriptional regulatory networks (TRNs) are pivotal in deciphering the molecular mechanisms underlying cancer progression. However, integrating multi-omics data to construct context-specific TRNs and dissecting their pan-cancer relevance remain challenging. Here, we developed a framework integrating RNA-seq, ATAC-seq, and DNA methylation data to reconstruct TRNs across 15 cancer types from The Cancer Genome Atlas (TCGA). Using the PECA model, we identified TF-RE-TG (transcription factor - regulatory element - target gene) regulatory relationships and refined networks using protein-protein interaction (PPI) data and methylation-aware motif filtering. Our analysis revealed cancer-specific and pan-cancer dysregulated TFs and TGs enriched in processes such as apoptosis, proliferation, and immune response. Notably, hub TFs like TBP and SP2 orchestrated pan-cancer modules linked to DNA repair and cell cycle regulation. Survival analysis identified prognostic TGs (e.g., ANLN, DOCK6), while enhancer and methylation analyses highlighted epigenetic drivers (e.g., SAPCD2, WRAP53) in tumorigenesis. Our study provides a comprehensive resource of cancer TRNs, unveiling conserved regulatory mechanisms and epigenetic vulnerabilities, with implications for precision oncology.