Phylogenetics and genomic variation of two genetically distinct Hepatocystis clades isolated from shotgun sequencing of wild primate hosts
Phylogenetics and genomic variation of two genetically distinct Hepatocystis clades isolated from shotgun sequencing of wild primate hosts
Haffener, P. E.; Hopson, H. D.; Leffler, E. M.
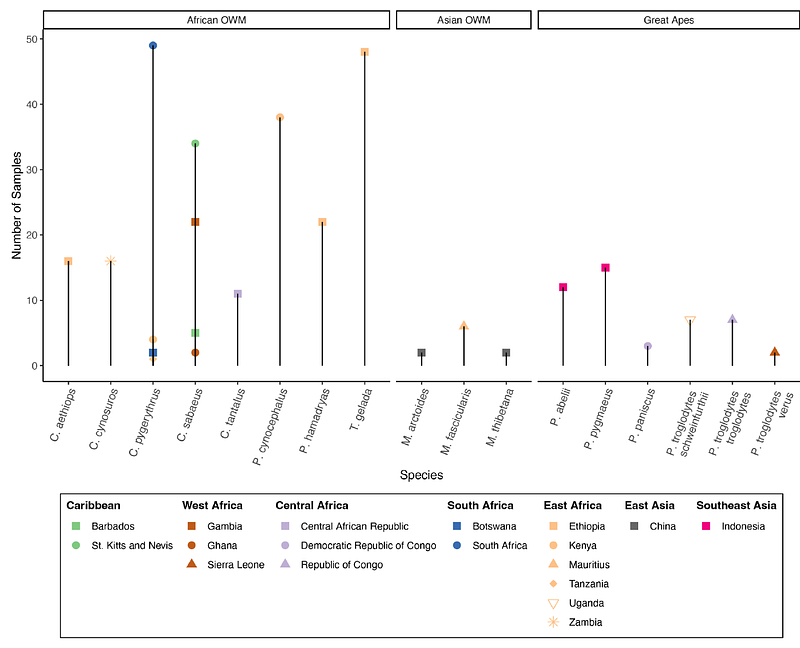
AbstractHepatocystis are apicomplexan parasites nested within the Plasmodium genus that infect primates and other vertebrates, yet few isolates have been genetically characterized. Using taxonomic classification and mapping characteristics, we searched for Hepatocystis infections within publicly available, blood-derived low coverage whole genome sequence (lcWGS) data from 326 wild non-human primates (NHPs) in 17 genera. We identified 30 Hepatocystis infections in Chlorocebus and Papio samples collected from locations in west, east, and south Africa. Hepatocystis cytb sequences from Papio hosts phylogenetically clustered with previously reported isolates from multiple NHP taxa whereas sequences from Chlorocebus hosts form a separate cluster, suggesting they represent a new host-specific clade of Hepatocystis. Additionally, there was no geographic clustering of Hepatocystis isolates suggesting both clades of Hepatocystis could be found in NHPs throughout sub-Saharan Africa. Across the genome, windows of high SNP density revealed candidate hypervariable loci including Hepatocystis-specific gene families possibly involved in immune evasion and genes that may be involved in adaptation to their insect vector and hepatocyte invasion. Overall, this work demonstrates how lcWGS data from wild NHPs can be leveraged to study the evolution of apicomplexan parasites and potentially test for association between host genetic variation and parasite infection.


