Quality Matters: Deep Learning-Based Analysis of Protein-Ligand Interactions with Focus on Avoiding Bias
Quality Matters: Deep Learning-Based Analysis of Protein-Ligand Interactions with Focus on Avoiding Bias
Sellner, M. S.; Lill, M. A.; Smiesko, M.
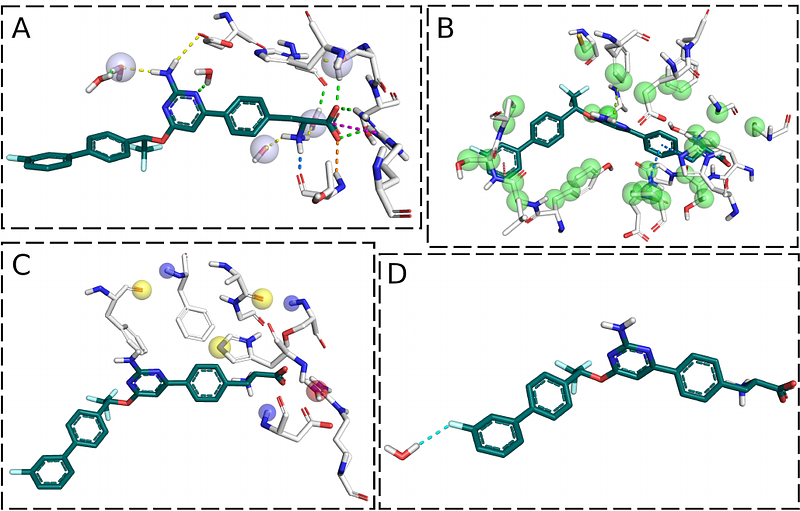
AbstractThe efficient and accurate prediction of protein-ligand binding affinities is an extremely appealing yet still unresolved goal in computational pharmacy. In recent years, many scientists have taken advantage of the remarkable progress of deep learning and applied it to address this issue. Despite all the advances in this field, there is increasing evidence that the typically applied validation of these methods is not suitable for medicinal chemistry applications. This work assesses the importance of dataset quality and proper dataset splitting techniques demonstrated on the example of the PDBbind dataset. We also introduce a new tool for the analysis of protein-ligand complexes, called po-sco. Po-sco allows the extraction of interaction information with much higher detail and comprehensibility than the tools available to date. We trained a transformer-based deep learning model to generate protein-ligand interaction fingerprints that can be utilized for downstream predictions, such as binding affinity. When using po-sco, this model generated predictions that were superior to those based on commonly used PLIP and ProLIF tools. We also demonstrate that the quality of the dataset is more important than the number of data points and that suboptimal dataset splitting can lead to a significant overestimation of model performance.