BRIDGE: A Coarse-Grained Architecture to Embed Protein-Protein Interactions for Therapeutic Applications
BRIDGE: A Coarse-Grained Architecture to Embed Protein-Protein Interactions for Therapeutic Applications
Khade, P. M.; Watkins, A. M.
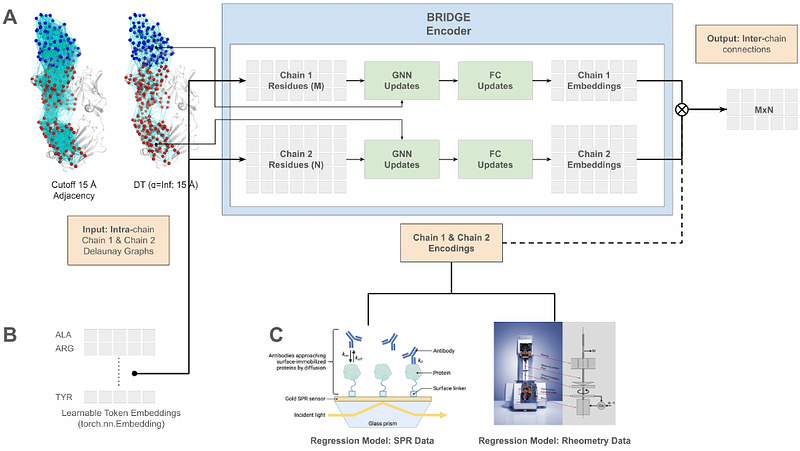
AbstractIn this work, we present Biophysical Representation of Interfaces via Delaunay-based Graph Embeddings (BRIDGE), a coarse-grained graph neural network that captures embeddings containing meaningful information about protein-protein interactions. The BRIDGE model takes as input graphs defined by the Delaunay tesselation of the individual chains and is pre-trained to predict the Delaunay adjacency at the protein-protein interface. The model achieves state-of-the-art performance in this task. The biophysical information captured by the BRIDGE embedding layer due to this pre-training task can further be used for downstream tasks, including for therapeutically relevant property prediction. We demonstrate the use of these embeddings by training models to predict antibody-antigen (Ab-Ag) affinity and antibody viscosity. These predictors, which require only the structures of the individual molecules in isolation, are especially well suited to large molecule therapeutic applications where complex structures are rarely determined until the very latest stages of a project.