Environmental RNA/DNA Metabarcoding for Ecological Risk Assessment of Chemicals Based on Response of Benthic Communities in Natural Environments
Environmental RNA/DNA Metabarcoding for Ecological Risk Assessment of Chemicals Based on Response of Benthic Communities in Natural Environments
Inoue, Y.; Miyata, K.; Yamane, M.; Honda, H.
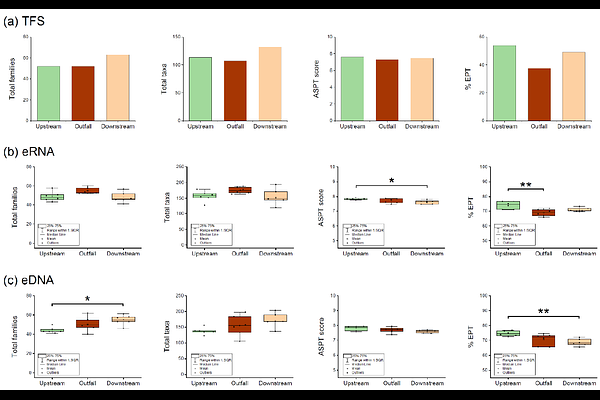
AbstractTo achieve the current Nature Positive goals, it is important to identify the effects of chemicals on ecosystems and strategically reduce their risks. We therefore estimated the safe concentration (SC) of linear alkylbenzene sulfonate (LAS) by investigating the relationship between benthic communities detected via environmental RNA (eRNA) and environmental DNA (eDNA) metabarcoding and the water quality in a river contaminated with LAS. Non-metric multidimensional scaling of benthic communities detected via eRNA indicated significant differences between contaminated and non-contaminated sites, and redundancy analysis showed significant correlation between the benthic communities and LAS concentration. These results demonstrate that eRNA is more sensitive than eDNA for capturing the community changes that result from LAS contamination. No significant differences were observed in biodiversity indices between the highest LAS concentration site and non-contaminated site; thus, the SC was estimated at >47 ug/L. The results suggest that, although hazard assessment using the assessment factor (AF) leads to conservative estimation of the effects of chemicals because the predicted no-effect concentration in previous studies is lower than the SC, hazard assessment based on the response of biological communities using eRNA metabarcoding can refine such ecological assessments, developing the environmental management standards to achieve nature-positive goals.