Elucidating DNA-binding protein dynamics in Salmonella Typhimurium within macrophages using a breakthrough low-input ChIP-exo approach
Elucidating DNA-binding protein dynamics in Salmonella Typhimurium within macrophages using a breakthrough low-input ChIP-exo approach
Park, J. Y.; Jang, M.; Choi, E.; Lee, S.-M.; Bang, I.; Woo, J.; Kim, S.; Lee, E.-J.; Kim, D.
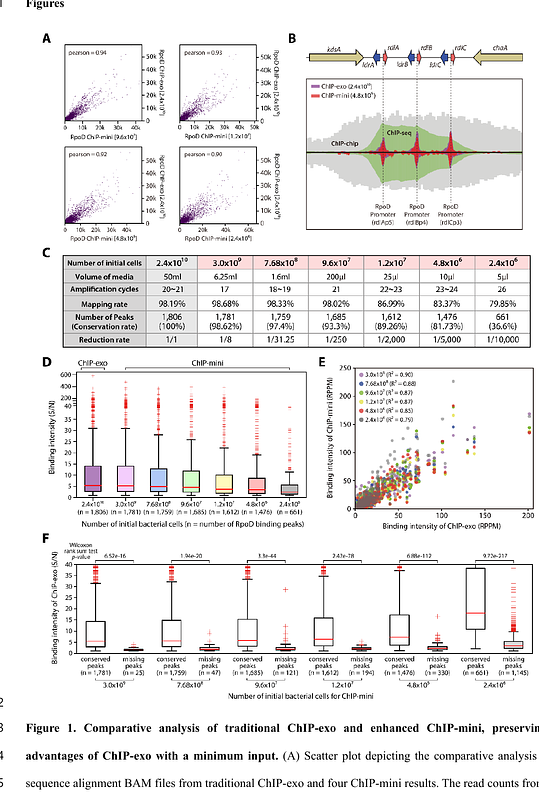
AbstractGenome-wide identification of binding profiles for DNA-binding proteins in Salmonella Typhimurium within macrophages is crucial for understanding virulence gene expression and cellular processes. However, this task remains challenging due to the limited amount of intracellular bacterial cells. Here, we present ChIP-mini, a low-input ChIP-exo utilizing 5,000-fold reduced number of initial bacterial cells and analysis pipeline (DiffExo), to identify genome-wide binding dynamics of DNA-binding proteins in host-infected pathogens. Applying ChIP-mini to intracellular S. Typhimurium, we identified 642 and 1,837 binding sites of H-NS and RpoD, respectively, with near-base pair resolution, elucidating their roles in transcriptional initiation. Post-infection, we observed 21 significant reductions in H-NS binding at intergenic regions, exposing the promoter region of virulence genes, such that those in Salmonella pathogenicity islands-2, 3 and effectors, facilitating RpoD binding for transcription initiation. Furthermore, we identified 24 novel RpoD binding sites and 19 significantly increased RpoD bindings at the transcription start sites of virulence genes. These findings substantially enhance our understanding of how H-NS and RpoD simultaneously coordinate the transcription initiation of virulence genes within macrophages. Collectively, this optimized method demonstrates a tool that can be broadly adapted to elucidate transcriptional regulatory networks of host-infected pathogens, revealing critical interactions between host and microbe.


