Recurrent disruption of tumour suppressor genes in cancer by somatic mutations in cleavage and polyadenylation signals
Recurrent disruption of tumour suppressor genes in cancer by somatic mutations in cleavage and polyadenylation signals
Kainov, Y.; Hamid, F.; Makeyev, E. V.
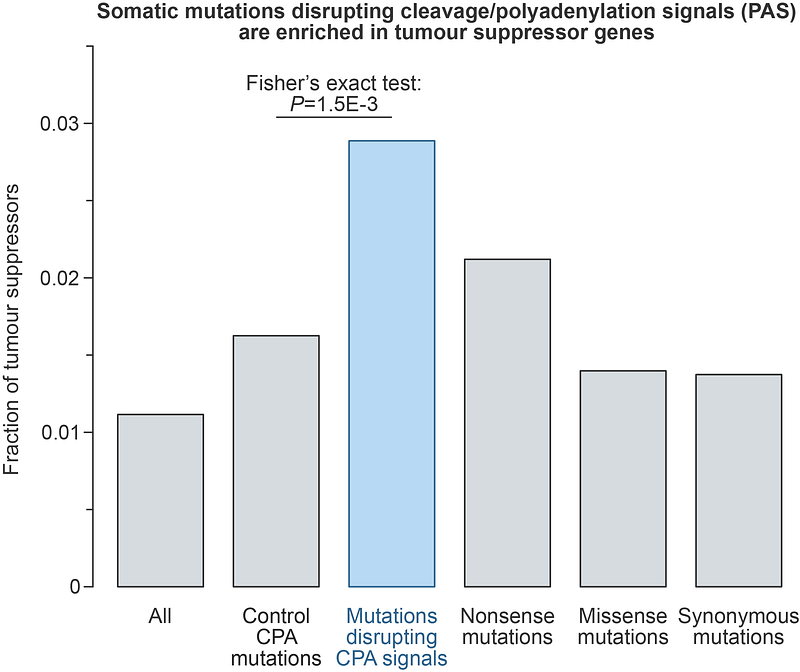
AbstractThe expression of eukaryotic genes relies on the precise 3\'-terminal cleavage and polyadenylation of newly synthesized pre-mRNA transcripts. Defects in these processes have been associated with various diseases, including cancer. While cancer-focused sequencing studies have identified numerous driver mutations in protein-coding sequences, noncoding drivers - particularly those affecting the cis-elements required for pre-mRNA cleavage and polyadenylation - have received less attention. Here, we systematically analysed cancer somatic mutations affecting 3\'UTR polyadenylation signals using the Pan-Cancer Analysis of Whole Genomes (PCAWG) dataset. We found a striking enrichment of cancer-specific somatic mutations that disrupt strong and evolutionarily conserved cleavage and polyadenylation signals within tumour suppressor genes. Further bioinformatics and experimental analyses conducted as a part of our study suggest that these mutations have a profound capacity to downregulate the expression of tumour suppressor genes. Thus, this work uncovers a novel class of noncoding somatic mutations with significant potential to drive cancer progression.


