DARDN: Identifying transcription factor binding motifs from long DNA sequences using multi-CNNs and DeepLIFT
DARDN: Identifying transcription factor binding motifs from long DNA sequences using multi-CNNs and DeepLIFT
Cho, H. J.; Wang, Z.; Cong, Y.; Bekiranov, S.; Zhang, A.; Zang, C.
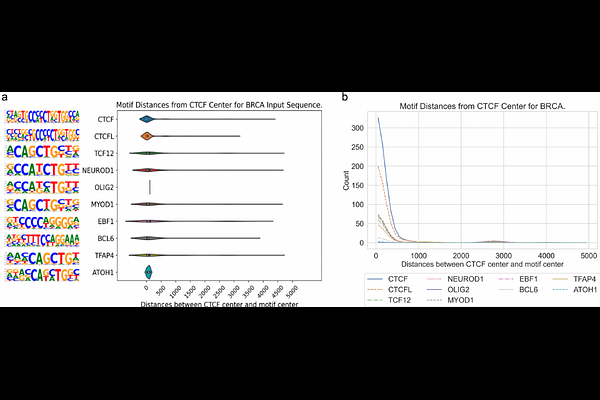
AbstractMotivation Characterization of regulatory elements in DNA sequence is a key task in functional genomics. CTCF exhibits specific binding patterns in the genome of cancer cells and has a non-canonical function to facilitate oncogenic transcription program by cooperating with transcription factors bound at flanking distal regions. Identification of sequence motifs from a broad genomic region surrounding cancer-specific CTCF binding sites can help find active transcription factors in a cancer type. However, the long DNA sequences without localization information makes it difficult to perform conventional motif enrichment analysis. Results We present DNAResDualNet (DARDN), a computational method that utilizes convolutional neural networks (CNNs) coupled with feature discovery using DeepLIFT, for identifying DNA sequence features that can differentiate two sets of lengthy DNA sequences. Evaluation on DNA sequences associated with CTCF binding sites in T-cell acute lymphoblastic leukemia (T-ALL) and other cancer types demonstrates DARDN\'s ability in classifying DNA sequences surrounding cancer-specific CTCF binding from control constitutive CTCF binding and identifying sequence motifs for transcription factors potentially active in each specific cancer type. We identified motifs for potential oncogenic transcription factors in T-ALL, acute myeloid leukemia (AML), breast cancer (BRCA), colorectal cancer (CRC), lung adenocarcinoma (LUAD), and prostate cancer (PRAD). Our work demonstrates the power of advanced machine learning and feature discovery approach in finding biologically meaningful information from complex DNA sequence data.