Intact protein barcoding enables one-shot identification of CRISPRi strains and their metabolic state
Intact protein barcoding enables one-shot identification of CRISPRi strains and their metabolic state
Pahl, V.; Lubrano, P.; Petras, D.; Link, H.
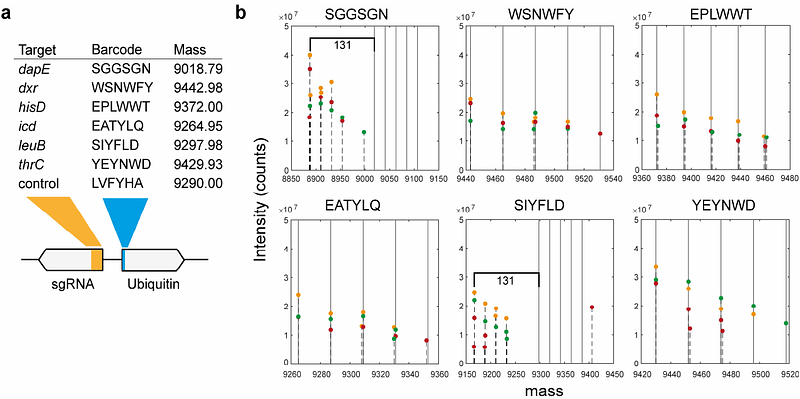
AbstractSimultaneous measurements of library barcodes and metabolites can facilitate the screening of metabolically engineered bacterial libraries. Here, we introduce a scalable method for the parallel measurement of protein-based library barcodes and metabolites using flow-injection mass spectrometry (FI-MS). Protein barcodes are based on ubiquitin, a small 76-amino-acid protein, which we labeled with N-terminal tags of six amino acids to identify bacterial strains. We demonstrate that FI-MS effectively detects intact ubiquitin proteins and identifies the N-terminal barcodes. In the same analysis, we semi-quantify relative concentrations of primary metabolites. As a proof of concept, we engineered six ubiquitin-barcoded CRISPRi strains targeting metabolic enzymes, and analyzed their metabolic profiles and ubiquitin barcodes using FI-MS. Our results show distinct metabolome changes within metabolic pathways targeted by CRISPRi. This method enables fast and simultaneously detection of library barcodes and intracellular metabolites, opening up new possibilities to measure metabolomes of engineered bacteria at scale.


