Discover the Maze-like Pathway for Glabridin Biosynthesis
Discover the Maze-like Pathway for Glabridin Biosynthesis
Zhang, Z.; Li, W.; Meng, F.; Jin, Y.; Sun, W.; Kuang, Q.; Ren, S.; Liu, W.; Zhang, L.; Qin, L.; Lv, B.; Jia, H.; Li, C.
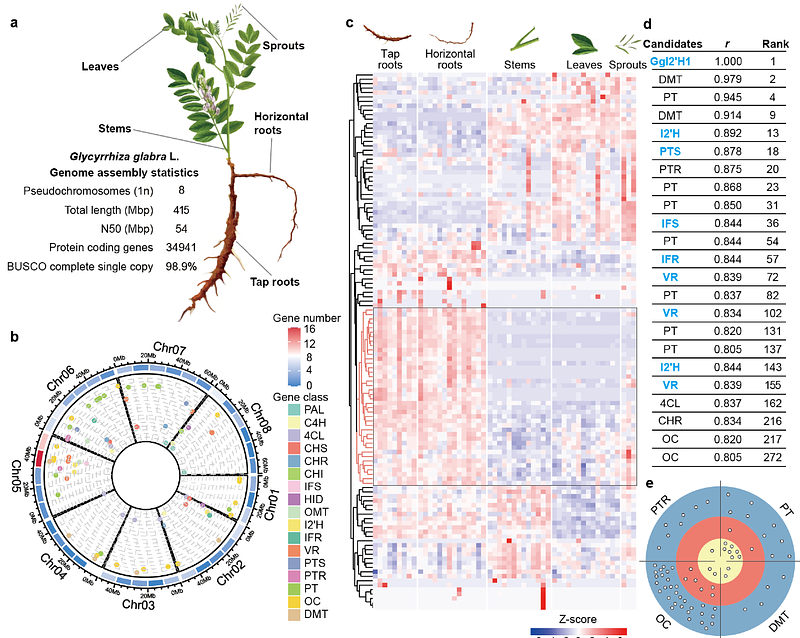
AbstractTailoring enzymes diversify plant metabolite scaffolds through complex, context-dependent modifications, generating maze-like biosynthetic networks that complicate metabolic pathway reconstruction. Here, we systematically deconstructed and reconstructed the biosynthetic network of glabridin, a valuable skin-whitening isoflavone from Glycyrrhiza. By integrating metabolic pathway mining with genome and 183 transcriptomes, we identified four functional routes among sixteen theoretical possibilities and uncovered a previously uncharacterized, ladder-like multi-route tailoring network. Reconstruction of such architecture in yeast revealed metabolic redundancy and interconnectivity confer unexpected robustness, enabling higher production efficiencies compared to a single-route design. Further modular engineering enabled the de novo biosynthesis of glabridin in yeast. Our work establishes a generalizable framework for reconstructing metabolic mazes and demonstrates that multi-route architectures can be harnessed to enhance the robustness and productivity of cell factories.