scMILD: Single-cell Multiple Instance Learning for Sample Classification and Associated Subpopulation Discovery
scMILD: Single-cell Multiple Instance Learning for Sample Classification and Associated Subpopulation Discovery
Jeong, K.; Choi, J.; Kim, K.
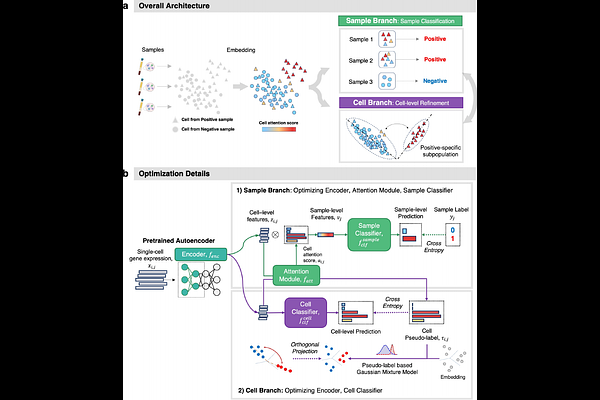
AbstractSingle-cell transcriptomics enables the study of cellular heterogeneity, but current unsupervised strategies make it challenging to associate individual cells with sample conditions. We propose scMILD, a weakly supervised learning framework based on Multiple Instance Learning, which leverages sample-level labels to identify condition-associated cell subpopulations. scMILD employs a dual-branch architecture to perform sample-level classification and cell-level representation learning simultaneously. We validated the model\'s reliable identification of condition-associated cells using controlled simulation studies with CRISPR-perturbed cells. Evaluated on diverse single-cell RNA-seq datasets, including Lupus, COVID-19, and Ulcerative Colitis, scMILD consistently outperformed state-of-the-art models and identified condition-specific cell subpopulations consistent with the original studies\' findings. This demonstrates scMILD\'s potential for exploring cellular heterogeneity underlying various biological conditions and its applicability in different disease contexts.