DTMol: Pocket-based Molecular Docking using Diffusion Transformers
DTMol: Pocket-based Molecular Docking using Diffusion Transformers
Teng, H.; Wang, R.; Shen, Y.; Yuan, Y.; Kingsford, C.
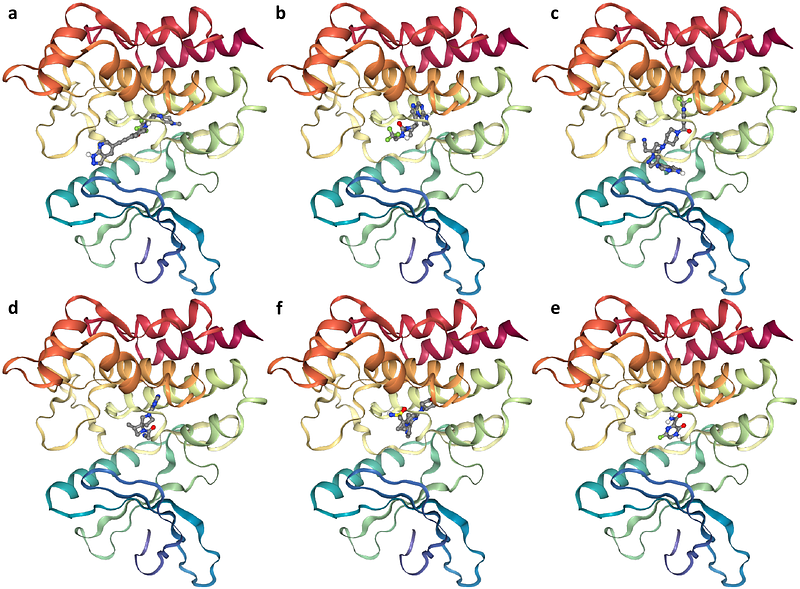
AbstractMolecular docking --- predicting the binding structure of a small molecule ligand to a protein --- is a crucial task in computational chemistry and drug discovery. Traditional docking methods relying on scoring functions tend to be slow and inaccurate. Recent deep learning methods, especially diffusion-based generative models, have significantly improved the accuracy and computational efficiency of molecular docking. However, these methods still face challenges, particularly in the pocket-based docking setting, which involves docking a ligand when a protein pocket structure --- a cavity of the protein with potential ligand-binding capabilities --- is given. We introduce DTMol, a novel generative deep learning model designed to tackle the pocket-based molecular docking problem. Our model integrates a pretrained molecular representation framework with a new SE(3)-equivariant diffusion transformer architecture. The pretrained framework generates representations of both protein pockets and ligands, while the diffusion transformer effectively captures interaction information between them. Testing on the PDB-Bind dataset demonstrates that our method outperforms traditional docking methods and deep learning-based baselines. The efficacy of DTMol is further validated through a virtual screening task targeting Janus kinase 2 (PDB ID: 6BBV), followed by experimental validation of the top-ranked compounds via a protein kinase activity assay.