Improving bacterial metagenomic research through long read sequencing
Improving bacterial metagenomic research through long read sequencing
Greenman, N. A.; Hassouneh, S. A.-D.; Johnston, C.; Azarian, T.
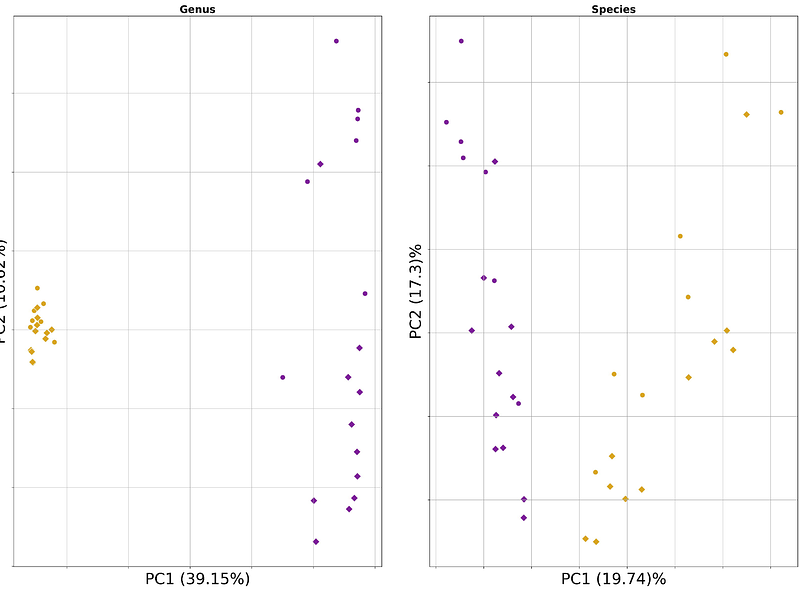
AbstractMetagenomic sequencing analysis is central to investigating microbial communities in clinical and environmental studies. Short read sequencing remains the primary data type for metagenomic research. However, long read sequencing promises the advantages of improved metagenomic assembly and resolved taxonomic identification. To assess the comparative performance of short and long read sequencing data for metagenomic analysis, we simulated short and long read datasets using increasingly complex metagenomes of 10, 20, and 50 microbial taxa. In addition, an empirical dataset of paired short and long read data from mouse fecal pellets was generated to assess feasibility. We compared metagenomic assembly quality, taxonomic classification capabilities, and metagenome-assembled genome recovery rates for both simulated and real metagenomic sequence data. We show that long read sequencing data significantly outperforms short read sequencing. For simulated long read datasets, metagenomic assemblies were completer and more contiguous with higher rates of metagenome-assembled genome recovery. This resulted in more precise taxonomic classifications and increased accuracy of relative abundance estimates. Analysis of empirical data demonstrated that sequencing technology directly affects compositional results. Overall, we highlight strengths of long read sequencing for metagenomic research of microbial communities over traditional short read approaches.