Single cell spatial proteomics maps human liver zonation patterns and their vulnerability to fibrosis
Single cell spatial proteomics maps human liver zonation patterns and their vulnerability to fibrosis
Weiss, C. A.; Brown, L. A.; Miranda, L.; Pellizzoni, P.; Ben-Moshe, S.; Steigerwald, S.; Remmert, K.; Hernandez, J.; Bogwardt, K.; Rosenberger, F. A.; Porat-Shliom, N.; Mann, M.
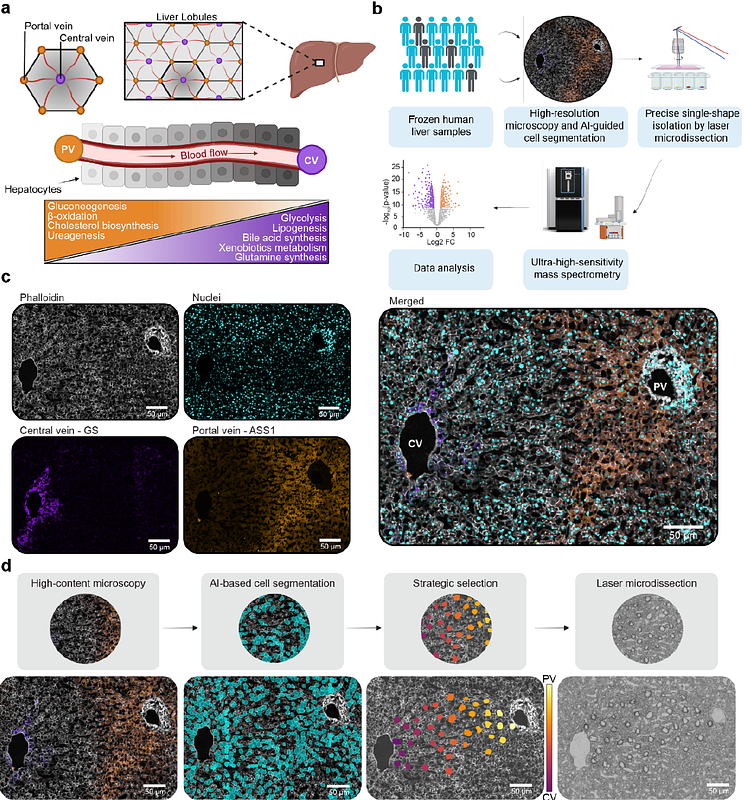
AbstractUnderstanding protein distribution patterns across tissue architecture is crucial for deciphering organ function in health and disease. Here, we applied single-cell Deep Visual Proteomics to perform spatially-resolved proteome analysis of individual cells in native tissue. We combined this with a novel strategic cell selection pipeline and a continuous protein gradient mapping framework to investigate larger clinical cohorts. We generated a comprehensive spatial map of the human hepatic proteome by analyzing hundreds of individual hepatocytes from 18 individuals. Among more than 2,500 proteins per cell about half exhibited zonated expression patterns. Cross-species comparison with mouse data revealed conserved metabolic functions and human-specific features of liver zonation. Analysis of fibrotic samples demonstrated widespread disruption of protein zonation, with pericentral proteins being particularly susceptible. Our study provides a comprehensive resource of human liver organization while establishing a broadly applicable framework for spatial proteomics analyses along tissue gradients.