Modification of regulatory tyrosines biases human Hsp90α for interaction with cochaperones and clients
Modification of regulatory tyrosines biases human Hsp90α for interaction with cochaperones and clients
Huo, Y.; Karnawat, R.; Liu, L.; Kniess, R. A.; Gross, M.; Chen, X.; Mayer, M. P.
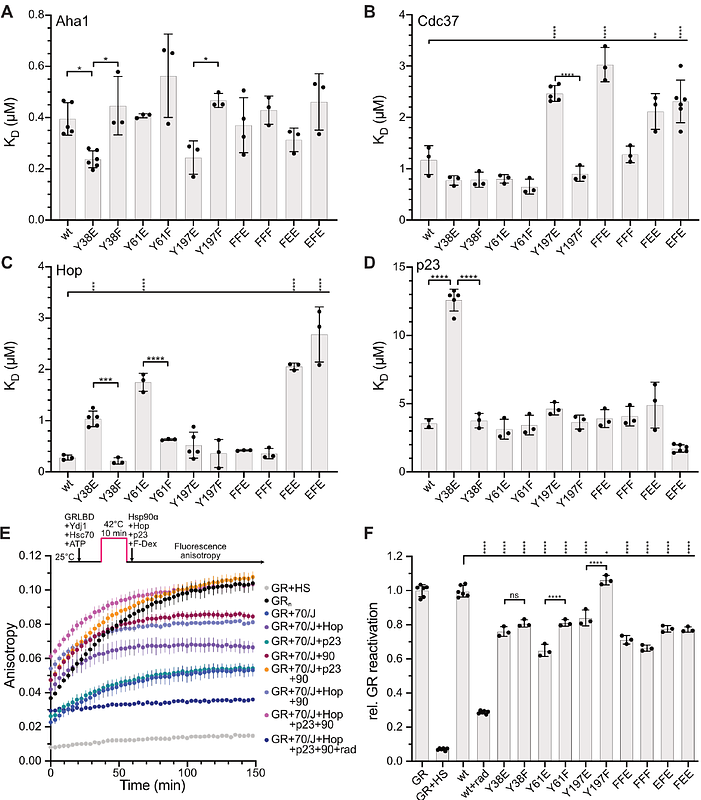
AbstractThe highly conserved Hsp90 chaperones control stability and activity of many essential signaling and regulatory proteins including many protein kinases, E3 ligases and transcription factors. Thereby, Hsp90s couple cellular homeostasis of the proteome to cell fate decisions. High-throughput mass spectrometry revealed 178 and 169 posttranslational modifications (PTMs) for human cytosolic Hsp90 and Hsp90{beta}, but for only a few of the modifications the physiological consequences are investigated in some detail. In this study, we explored the suitability of the yeast model system for the identification of key regulatory residues in human Hsp90. Replacement of three tyrosine residues known to be phosphorylated by phosphomimetic glutamate and by non-phosphorylatable phenylalanine individually and in combination influenced yeast growth and the maturation of 7 different Hsp90 clients in distinct ways. Furthermore, wild-type and mutant Hsp90 differed in their ability to stabilize known clients when expressed in HepG2 HSP90AA1-/- cells. The purified mutant proteins differed in their interaction with the cochaperones Aha1, Cdc37, Hop and p23 and in their support of the maturation of glucocorticoid receptor ligand binding domain in vitro. In vivo and in vitro data correspond well to each other confirming that the yeast system is suitable for the identification of key regulatory sites in human Hsp90s. Our findings indicate that even closely related clients are affected differently by the amino acid replacements in the investigated positions, suggesting that PTMs could bias Hsp90\'s client specificity.


