Introducing the Y chromosome ancestral reference sequence - Improving the capture of human evolutionary information
Introducing the Y chromosome ancestral reference sequence - Improving the capture of human evolutionary information
Koksal, Z.; Preussner, A.; Leinonen, J.; Tukiainen, T.
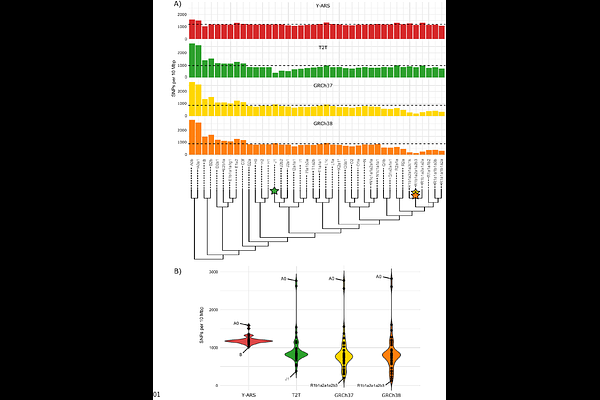
AbstractReference sequences are essential for reproducible genetic analyses but are often chosen without regard to evolutionary relevance within the analyzed species. The human Y chromosome (chrY) is widely used in evolutionary studies, yet current references represent evolutionarily young sequences, which can lead to misleading variant calling. To address this issue, we constructed a Y-chromosomal ancestral-like reference sequence (Y-ARS) to improve the detection of evolutionarily informative variants on the Y chromosome. The Y-ARS was constructed by applying a weighted maximum parsimony approach to human and primate Y chromosome sequences. To benchmark the performance of the Y-ARS, 40 chrY short-read sequences from diverse haplogroups were aligned to Y-ARS and existing references (GRCh37, GRCh38, and T2T-CHM13). Overall, the Y-ARS yielded the highest and most consistent number of SNPs per sample (mean=1197; SD=105), while other references yielded on average fewer variants (mean=866-968) and showed greater variability across samples (SD=457-531) depending on their phylogenetic distance from the reference. Additionally, alignments to the Y-ARS resulted in calling solely SNPs with evolutionarily derived alleles, while alignments to other references resulted in calling on average 44% SNPs with ancestral alleles. This study demonstrates how the existing reference sequences fail to capture the full range of evolutionary information on the chrY. The Y-ARS improves capturing evolutionary information on the chrY, making it a valuable resource for various evolutionary applications, such as TMRCA estimations and phylogenetic analyses. Finally, alongside the Y-ARS, we provide a publicly available tool, polaryzer, to annotate variants as ancestral or derived in pre-aligned chrY data.