Circos Plots for Genome Level Interpretation of Genomic Prediction Models
Circos Plots for Genome Level Interpretation of Genomic Prediction Models
Tomura, S.; Powell, O. M.; Wilkinson, M. J.; Cooper, M.
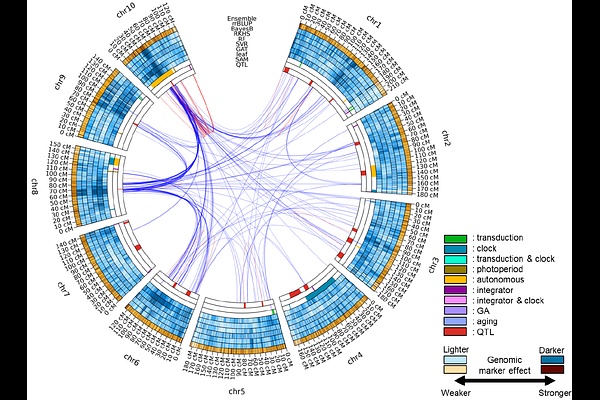
AbstractInvestigations of genomic prediction models have focused primarily on the level of metrics summarising the accuracy and error of predicted phenotypes. Analysis of the genomic regions contributing to the genomic prediction models can deepen our understanding of the trait genetic architecture. Here, we develop circos plots to visualise how different genomic prediction models quantify marker effects. The genetic architecture was inferred using interpretable genomic prediction models to characterise each method at the genome level. Application is demonstrated for the trait days to anthesis (DTA) in the TeoNAM dataset. The results indicate that genomic prediction models can capture different views of trait genetic architecture, even when their overall profiles of prediction accuracy are similar. Combinations of diverse views of the genetic architecture for the DTA trait in the TeoNAM study might explain the improved prediction performance achieved by ensembles of prediction models, aligned with the implication of the Diversity Prediction Theorem. In addition to identifying well-known genomic regions contributing to the genetic architecture of DTA in maize, the ensemble of genomic prediction models highlighted several new genomic regions that have not been previously identified for DTA. Finally, different views of trait genetic architecture were observed across populations, within the TeoNAM design, highlighting challenges for between-population genomic prediction. A deeper understanding of genomic prediction models with enhanced interpretability can reveal several critical findings at the genome level from the inferred genetic architecture, providing insights into the improvement of genomic prediction for crop breeding programs.