mt.surv: Multi-Threshold Survival Analysis for Associating Continuous Predictor Variables with Time-to-Event Outcomes
mt.surv: Multi-Threshold Survival Analysis for Associating Continuous Predictor Variables with Time-to-Event Outcomes
Loncar, A. J.; Hoyd, R.; Liu, Y.; Dravillas, C.; Dhrubo, D.; Spakowicz, D. J.
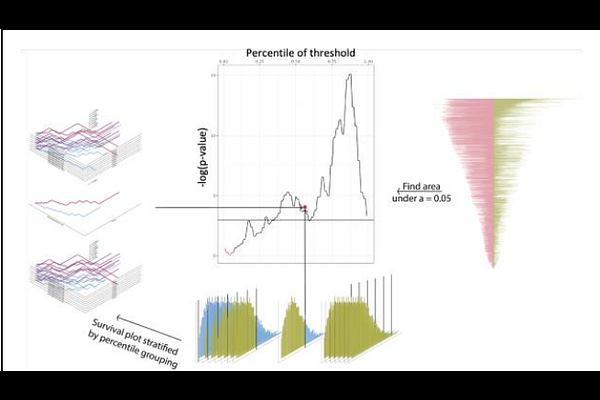
AbstractTime-to-event models are a useful and common approach to infer the importance of biological variables. Most often, the predictor variable is binarized and associated with censored time to death, a so-called Kaplan-Meier curve. However, the threshold to binarize the continuous variable is often arbitrary. We sought a rigorous way to define thresholds and evaluate the strength and consistency of association with the event of interest. We present {mt.surv}, an R package for multi-threshold survival analyses. The primary function performs a time-to-event analysis, where a continuous predictor variable is stratified into two groups at an arbitrary number of places, defined by the percentile of the distribution. The result can be visualized with a function that creates a custom line plot with the -log of a log-likelihood p-value against the threshold percentile. A third function operates on this plot and calculates the area above a significance threshold, creating a scalar that can be used to rank biological variables for their association with the event of interest. This framework can be broadly applied to any continuous predictor variable including, but not limited to, gene expression and microbial abundances. Several helper functions are included for structuring input data and job submission in a cluster framework. We found that this method has value in discovery-type analyses where one lacks prior information about appropriate stratification thresholds. However, we found additional biological in-sight is possible, indeed, quite common, as many variables show different associations at differ-ent stratification thresholds.