Single-chain permuted proteins for dimerization-based control of protein activity and cellular processes
Single-chain permuted proteins for dimerization-based control of protein activity and cellular processes
Zeleznik Ramuta, T.; Snoj, J.; Kos, T.; Ljubetic, A.; Hafner-Bratkovic, I.; Jerala, R.
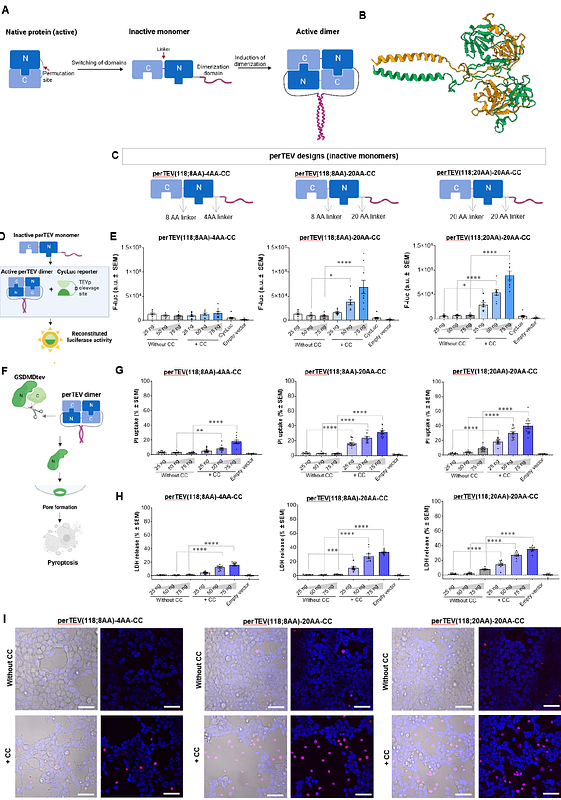
AbstractStrategies for detecting and controlling protein interactions play a critical role in gaining insight into molecular mechanisms of biological processes and for the control of cellular processes. Conditional protein reconstitution allows control of the selected protein function based on the proximity, defined by the genetically fused domain pairs, which may be regulated by chemical or biological signals. This typically requires two protein components in a stoichiometric ratio, which increases the complexity and genetic footprint with split segments often being unstable and prone to aggregation. To overcome this limitation, we developed an approach based on a permuted protein reconstitution by conditional dimerization (PROPER). According to this strategy, the N- and C-terminal domains of selected proteins are swapped and a loop is replaced by a short linker that prevents the functionality of a monomeric protein, which reconstitutes only upon di- or oligomerization, controlled by a genetically fused domain that dimerizes by a chemical signal or senses a dimeric target. This design principle was demonstrated on three proteins with diverse functions: a protease, luciferase, and a cytokine. We demonstrate chemically and biologically inducible systems that enable controllable induction of cell death, virus detection, and immune cell stimulation. The PROPER platform expands the chemical biology toolbox, with the benefits of split proteins accomplished by a single component.