Inferring physical cell-cell communication networks from scRNAseq data using univariate linear models.
Inferring physical cell-cell communication networks from scRNAseq data using univariate linear models.
Hameed, S. A.; Iglesias-Martinez, L. F.; kolch, W.; Zhernovkov, V.
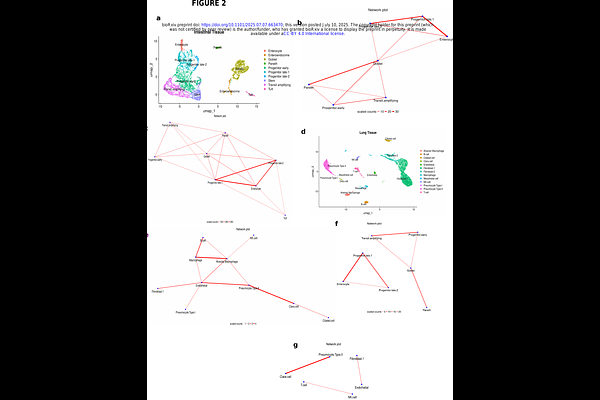
AbstractCells in tissues interact by direct physical contact or over short and long distances via secreted mediators. Cell-cell communication inference has now become routine in downstream scRNAseq analysis but this fails to capture physical cell-cell interactions due to tissue dissociation. Multiplets (mostly doublets) in scRNAseq may represent undissociated physically attached cells that become sequenced together. Hence, identifying multiplets may serve as a good starting point to harness scRNAseq data for physical cell-cell interaction inference. In this study, we develop a computational method which utilizes univariate linear models (ULM) to identify multiplets in scRNAseq datasets, predict their cellular compositions, and infer physical cell-cell interaction networks. Indeed, our method showed good sensitivity (~56%) with excellent precision (~99%) in predicting FACS sorted doublet cell pairs with known constituents (ground truth), recording comparable or superior performance over two other existing methods. Also, applying it to scRNAseq data of partially dissociated tissues containing real multiplets unraveled physical networks which recapitulated the microanatomical structures of the tested tissues. This further underscores the accuracy in our predictions to capture biologically meaningful interactions. Finally, we tested our method on classical scRNAseq datasets and obtained biologically reasonable results. For example, when tested on classical cancer scRNAseq datasets, we recovered important interactions which followed biologically plausible cell interactions, validated by cell-cell colocalization in matched spatial transcriptomics datasets. This reassured the accuracy of our method in depicting physical interactions only between cells that were truly in close proximity in tissues