MetaFX: feature extraction from whole-genome metagenomic sequencing data
MetaFX: feature extraction from whole-genome metagenomic sequencing data
Ivanov, A.; Popov, V.; Morozov, M.; Olekhnovich, E.; Ulyantsev, V.
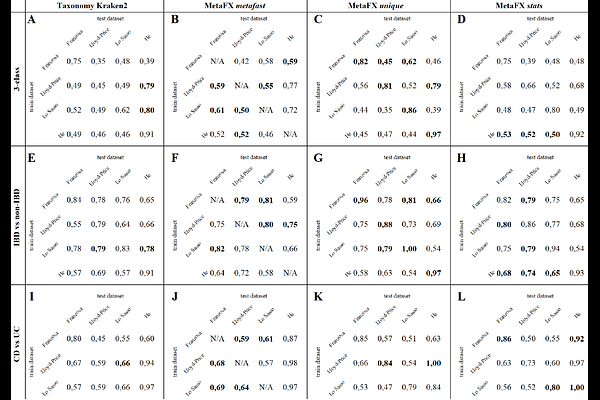
AbstractMotivation: Microbial communities consist of thousands of microorganisms and viruses and have a tight connection with an environment, such as gut microbiota modulation of host body metabolism. However, the direct relationship between the presence of certain microorganism and host state often remains unknown. Toolkits using reference-based approaches are limited to microbes present in databases. Reference-free methods often require enormous resources for metagenomic assembly or results in many poorly interpretable features based on k-mers. Results: Here we present MetaFX -- an open-source library for feature extraction from whole-genome metagenomic sequencing data and classification of groups of samples. Using a large volume of metagenomic samples deposited in databases, MetaFX compares samples grouped by metadata criteria (e.g. disease, treatment, etc) and constructs genomic features distinct for certain types of communities. Features constructed based on statistical k-mer analysis and de Bruijn graphs partition. Those features are used in machine learning models for classification of novel samples. Extracted features can be visualised on de Bruijn graphs and annotated for providing biological insights. We demonstrate the utility of MetaFX by building classification models for 590 human gut samples with inflammatory bowel disease. Our results outperform the previous research disease prediction accuracy up to 17%, and improves classification results compared to taxonomic analysis by 9 {+/-} 10% on average. Availability: MetaFX is a feature extraction toolkit applicable for metagenomic datasets analysis and samples classification. The source code, test data, and relevant information for MetaFX are freely accessible at https://github.com/ctlab/metafx under the MIT License.