Global and cross-omics feature aggregation improves single-cell multi-omics integration and clustering
Global and cross-omics feature aggregation improves single-cell multi-omics integration and clustering
guo, y.; chenyang, c.; zhu, z.; zhou, j.; Lu, H.; qian, y.; Liang, Z.
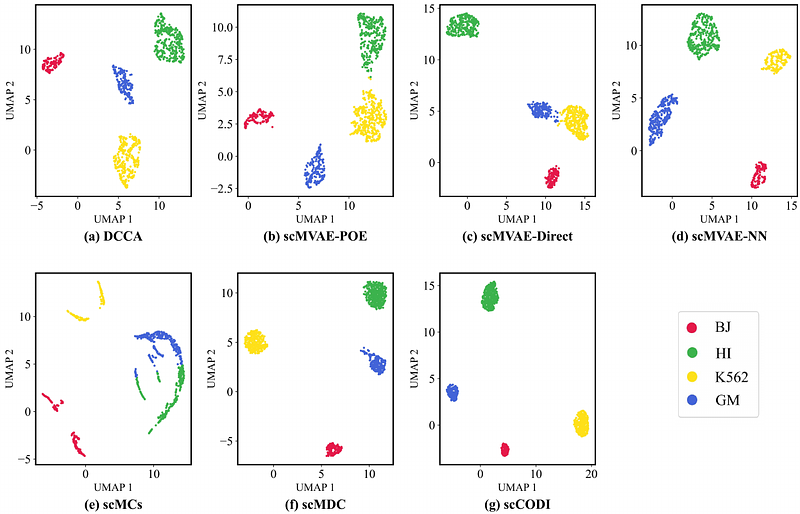
AbstractSequencing technology advancements enable the simultaneous measurement of multiple omics within single cells, thereby facilitating single-cell multi-omics analysis. Most existing deep multi-omics integration methods learn shared or omics-specific representations from multiple omics by omics-wise aggregation. However, these approaches often fail to account for the structural relationships among all cells. Here, we introduce scCODI, an innovative cross-omics deep learning model to integrate single-cell multi-omics data for cell clustering. This model processes paired omics datasets, combining single-cell transcriptomics with either epigenomics or proteomics data to facilitate a more comprehensive exploration of cellular heterogeneity. Specifically, the shared representation of two omics is derived through cross-omics feature aggregation, fully leveraging the complementarity of different omics within the same cell. Additionally, we align the omic-specific and shared representations of each cell using a global relationship-guided contrastive learning module, which enhances the similarity between the shared and omic-specific representations of the same cell. Extensive experiments demonstrate that scCODI outperforms previous methods in single-cell multi-omics data integration and cell clustering, as evidenced by its performance across nine paired multi-omics datasets. This makes it a promising tool for decoding complex biological mechanisms.